Search Count: 89
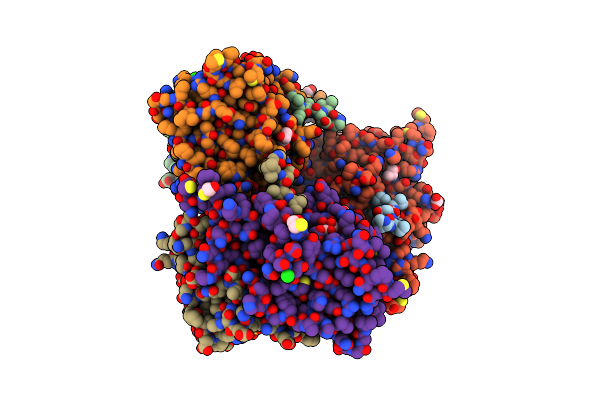 |
Tyrosine 447 Of Protocatechuate 3,4-Dioxygenase Controls Efficient Progress Through Catalysis
Organism: Pseudomonas putida
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2011-03-09 Classification: oxidoreductase/oxidoreductase Ligands: SO4, GOL, BME, TRS, CO3, FE, 4NC, CL |
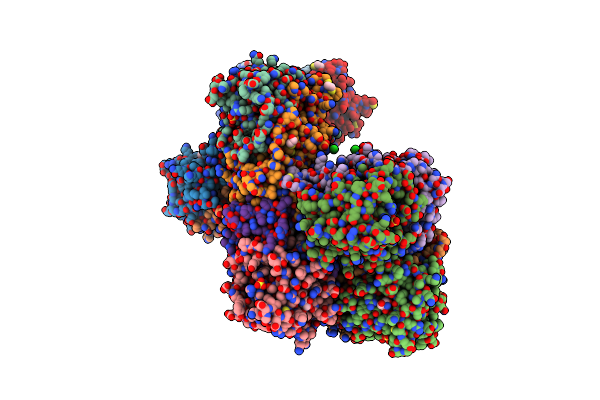 |
Tyrosine 447 Of Protocatechuate 3,4-Dioxygenase Controls Efficient Progress Through Catalysis
Organism: Pseudomonas putida
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2011-02-23 Classification: OXIDOREDUCTASE Ligands: SO4, BME, TRS, FE, CL, GOL |
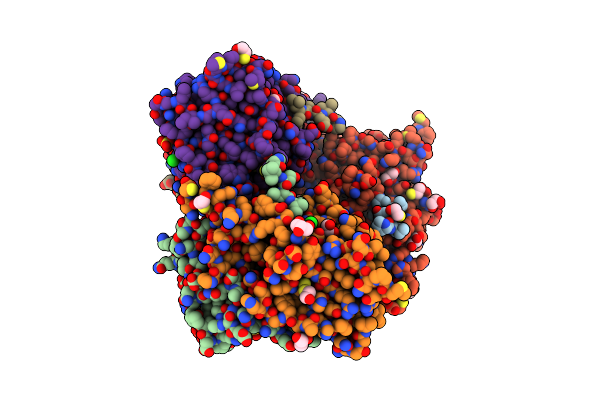 |
Tyrosine 447 Of Protocatechuate 34,-Dioxygenase Controls Efficient Progress Through Catalysis
Organism: Pseudomonas putida
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2011-02-16 Classification: OXIDOREDUCTASE Ligands: GOL, SO4, BME, CL, FE, DHB |
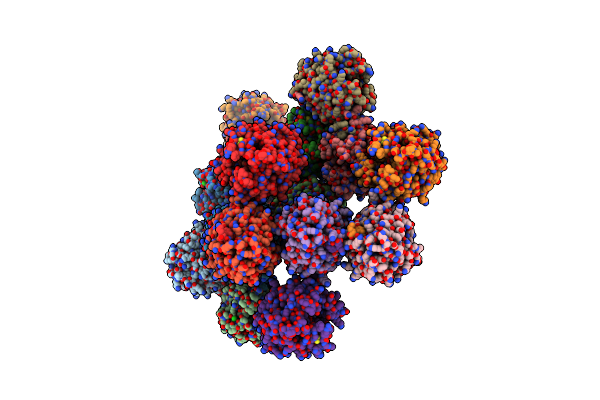 |
Structure Of Beta Hairpin Deletion Mutant Of Beta Toxin From Staphylococcus Aureus
Organism: Staphylococcus aureus
Method: X-RAY DIFFRACTION Resolution:3.35 Å Release Date: 2011-01-26 Classification: HYDROLASE Ligands: CL, NA |
 |
Crystal Structure Of Adipocyte Fatty Acid Binding Protein Covalently Modified With 4-Hydroxy-2-Nonenal
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:1.81 Å Release Date: 2010-08-25 Classification: LIPID BINDING PROTEIN Ligands: PO4 |
 |
Crystal Structure Of Adipocyte Fatty Acid Binding Protein Non-Covalently Modified With 4-Hydroxy-2-Nonenal
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2010-08-25 Classification: LIPID BINDING PROTEIN Ligands: CL, HNE, PO4 |
 |
Organism: Staphylococcus aureus
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2010-08-11 Classification: TOXIN Ligands: DGA |
 |
Crystal Structure Of Beta Toxin From Staphylococcus Aureus F277A, P278A Mutant
Organism: Staphylococcus aureus
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2010-07-14 Classification: TOXIN |
 |
Crystal Structure Of Beta Toxin From Staphylococcus Aureus F277A, P278A Mutant With Bound Calcium Ions
Organism: Staphylococcus aureus
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2010-07-14 Classification: TOXIN Ligands: CA, CL |
 |
Crystal Structure Of Beta Toxin From Staphylococcus Aureus F277A, P278A Mutant With Bound Magnesium Ions
Organism: Staphylococcus aureus rn4220
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2010-07-14 Classification: TOXIN Ligands: MG, PO4 |
 |
Organism: Staphylococcus aureus subsp. aureus
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2010-06-09 Classification: UNKNOWN FUNCTION Ligands: CL |
 |
Identification And Characterization Of A Small Molecule Inhibitor Of Fatty Acid Binding Proteins
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2009-09-22 Classification: PROTEIN BINDING Ligands: B64 |
 |
Organism: Staphylococcus aureus
Method: X-RAY DIFFRACTION Resolution:0.92 Å Release Date: 2009-06-30 Classification: TOXIN Ligands: ZN, IOD |
 |
Organism: Enterococcus faecalis
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2007-04-03 Classification: TRANSCRIPTION |
 |
Organism: Enterococcus faecalis
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2007-04-03 Classification: TRANSCRIPTION |
 |
Crystal Structure Of Wild-Type Protocatechuate 3,4-Dioxygenase From Acinetobacter Sp. Adp1
Organism: Acinetobacter sp.
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2006-09-05 Classification: OXIDOREDUCTASE Ligands: FE, OH |
 |
Crystal Structure Of Wild-Type Protocatechuate 3,4-Dioxygenase From Acinetobacter Sp. Adp1 In Complex With Catechol
Organism: Acinetobacter calcoaceticus
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2006-09-05 Classification: OXIDOREDUCTASE Ligands: FE, CAQ |
 |
Crystal Structure Of Wild-Type Protocatechuate 3,4-Dioxygenase From Acinetobacter Sp. Adp1 In Complex With 4-Hydroxybenzoate
Organism: Acinetobacter calcoaceticus
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2006-09-05 Classification: OXIDOREDUCTASE Ligands: FE, PHB |
 |
Crystal Structure Of Protocatechuate 3,4-Dioxygenase From Acinetobacter Sp. Adp1 Mutant R457S - Apo
Organism: Acinetobacter calcoaceticus
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2006-09-05 Classification: OXIDOREDUCTASE Ligands: FE, OH |
 |
Crystal Structure Of Protocatechuate 3,4-Dioxygenase From Acinetobacter Sp. Adp1 Mutant R457S In Complex With Protocatechuate
Organism: Acinetobacter calcoaceticus
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2006-09-04 Classification: OXIDOREDUCTASE Ligands: FE, DHB |

