Search Count: 25
 |
The Structure Of Ente With 3-(Prop-2-Yn-1-Yloxy)Benzoic Acid Sulfamoyl Adenosine
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.65 Å Release Date: 2024-07-31 Classification: LIGASE Ligands: VPT |
 |
The Structure Of Ente With2-Methyl-3-Chloro-Benzoic Acid Sulfamoyl Adenosine
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2024-07-31 Classification: LIGASE Ligands: VQ5 |
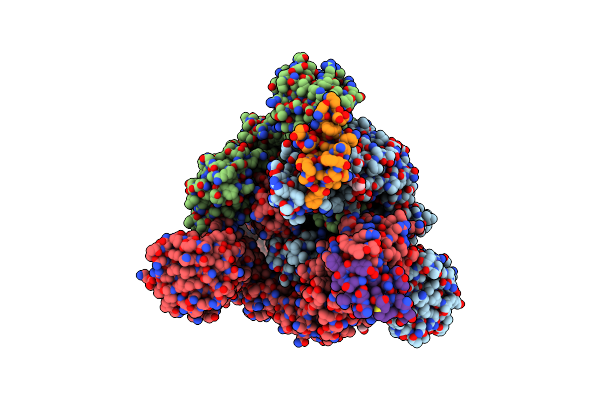 |
Conformation 1 Of Sars-Cov-2 Omicron Ba.1 Variant Spike Protein Complexed With Mo1 Fab
Organism: Severe acute respiratory syndrome coronavirus 2, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2023-05-10 Classification: VIRAL PROTEIN Ligands: NAG |
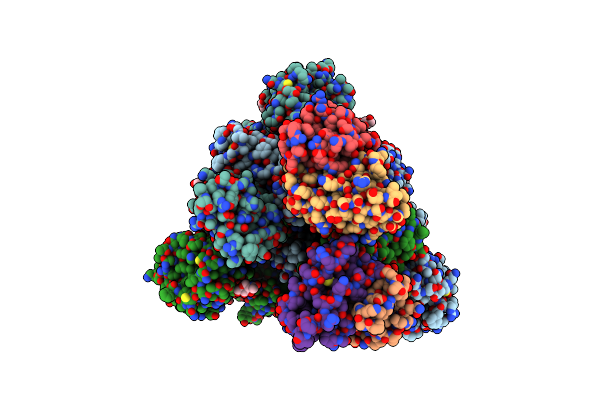 |
Conformation 2 Of Sars-Cov-2 Omicron Ba.1 Variant Spike Protein Complexed With Mo1 Fab
Organism: Severe acute respiratory syndrome coronavirus 2, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2023-05-10 Classification: VIRAL PROTEIN Ligands: NAG |
 |
Organism: Lama glama, Aquifex aeolicus
Method: X-RAY DIFFRACTION Resolution:3.65 Å Release Date: 2023-03-29 Classification: MEMBRANE PROTEIN/Transferase Ligands: P5L |
 |
In Meso Full-Length Rat Kmo In Complex With A Pyrazoyl Benzoic Acid Inhibitor
Organism: Rattus norvegicus
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2020-12-23 Classification: MEMBRANE PROTEIN Ligands: FAD, EGO, SO4, CL |
 |
In Meso Full-Length Rat Kmo In Complex With An Inhibitor Identified Via Dna-Encoded Chemical Library Screening
Organism: Rattus norvegicus
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2020-12-23 Classification: MEMBRANE PROTEIN Ligands: FAD, SO4, CL, EGU |
 |
Organism: Saccharomyces cerevisiae s288c
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2016-12-07 Classification: REPLICATION |
 |
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2013-04-10 Classification: DNA BINDING PROTEIN Ligands: EDO |
 |
Crystal Structure Of A 2-Fluoroxylotriosyl Complex Of The Vibrio Sp. Ax-4 Beta-1,3-Xylanase
Organism: Vibrio
Method: X-RAY DIFFRACTION Resolution:1.20 Å Release Date: 2013-03-06 Classification: HYDROLASE Ligands: DNX |
 |
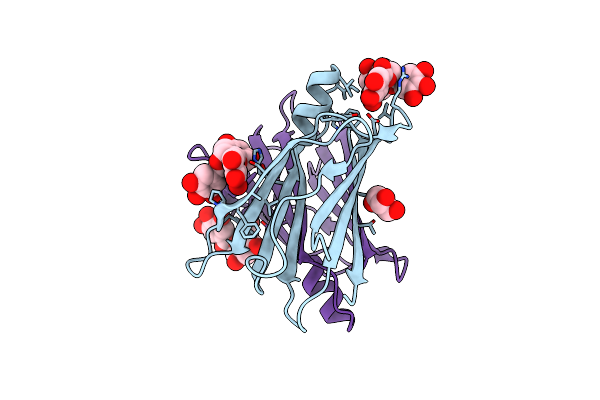
Crystal Structure Of Human Gluk2 Ligand-Binding Core In Complex With Novel Marine-Derived Toxins, Neodysiherbaine A
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2011-10-26 Classification: MEMBRANE PROTEIN Ligands: NDZ |
 |
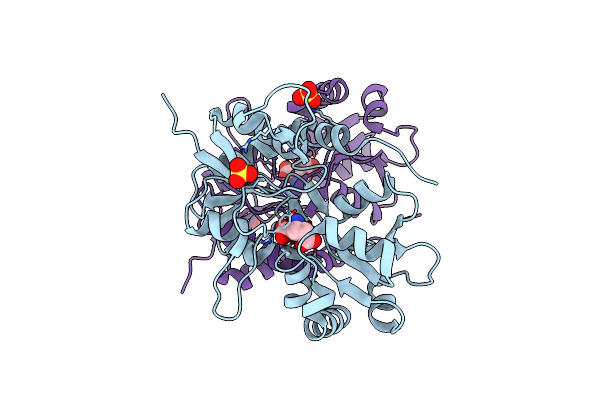
Crystal Structure Of V30M Transthyretin Complexed With (-)-Epigallocatechin Gallate (Egcg)
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2010-07-07 Classification: TRANSPORT PROTEIN Ligands: KDH, GOL |
 |
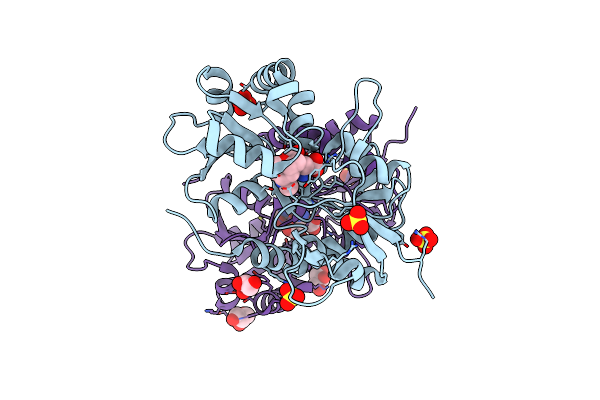
Crystal Structure Of The Human Glutamate Receptor, Glur5, Ligand-Binding Core In Complex With L-Glutamate In Space Group P1
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2010-01-19 Classification: MEMBRANE PROTEIN Ligands: GLU, SO4 |
 |
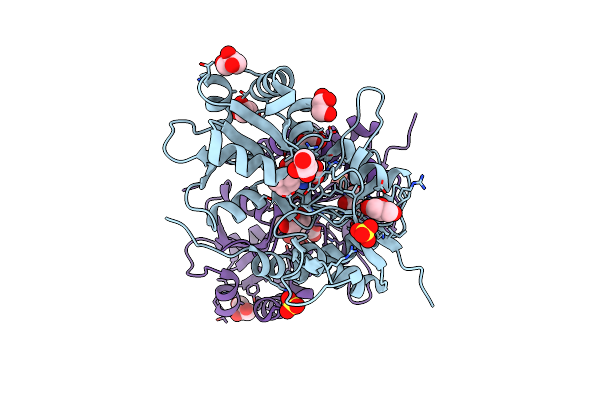
Crystal Structure Of The Human Glutamate Receptor, Glur5, Ligand-Binding Core In Complex With Dysiherbaine In Space Group P1
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2010-01-19 Classification: MEMBRANE PROTEIN Ligands: DYH, SO4, GOL |
 |
Crystal Structure Of The Human Glutamate Receptor, Glur5, Ligand-Binding Core In Complex With Neodysiherbaine A In Space Group P1
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2010-01-19 Classification: MEMBRANE PROTEIN Ligands: GOL, NDZ, SO4 |
 |
Crystal Structure Of The Human Glutamate Receptor, Glur5, Ligand-Binding Core In Complex With Msviii-19 In Space Group P1
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2010-01-19 Classification: MEMBRANE PROTEIN Ligands: GOL, MS8, SO4 |
 |
Crystal Structure Of The Human Glutamate Receptor, Glur5, Ligand-Binding Core In Complex With 8-Deoxy-Neodysiherbaine A In Space Group P1
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2010-01-19 Classification: MEMBRANE PROTEIN Ligands: GOL, 8DX, SO4 |
 |
Crystal Structure Of The Human Glutamate Receptor, Glur5, Ligand-Binding Core In Complex With 9-Deoxy-Neodysiherbaine A In Space Group P1
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2010-01-19 Classification: MEMBRANE PROTEIN Ligands: GOL, 9DX, SO4 |
 |
Crystal Structure Of The Human Glutamate Receptor, Glur5, Ligand-Binding Core In Complex With 8-Epi-Neodysiherbaine A In Space Group P1
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2010-01-19 Classification: MEMBRANE PROTEIN Ligands: GOL, 8EP, SO4 |
 |
Crystal Structure Of The Ligand-Binding Core Of The Human Ionotropic Glutamate Receptor, Glur5, In Complex With Glutamate
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2009-05-05 Classification: MEMBRANE PROTEIN Ligands: GLU |

