Search Count: 26
 |
Organism: Kluyveromyces lactis (strain atcc 8585 / cbs 2359 / dsm 70799 / nbrc 1267 / nrrl y-1140 / wm37)
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2022-09-14 Classification: TRANSFERASE Ligands: ZN |
 |
Organism: Kluyveromyces lactis (strain atcc 8585 / cbs 2359 / dsm 70799 / nbrc 1267 / nrrl y-1140 / wm37)
Method: X-RAY DIFFRACTION Resolution:2.19 Å Release Date: 2022-09-14 Classification: TRANSFERASE Ligands: ZN |
 |
Organism: Kluyveromyces lactis (strain atcc 8585 / cbs 2359 / dsm 70799 / nbrc 1267 / nrrl y-1140 / wm37)
Method: X-RAY DIFFRACTION Resolution:1.76 Å Release Date: 2022-09-14 Classification: TRANSFERASE Ligands: ZN |
 |
Organism: Kluyveromyces lactis (strain atcc 8585 / cbs 2359 / dsm 70799 / nbrc 1267 / nrrl y-1140 / wm37)
Method: X-RAY DIFFRACTION Resolution:1.51 Å Release Date: 2022-09-14 Classification: TRANSFERASE Ligands: ZN |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2020-04-29 Classification: STRUCTURAL PROTEIN |
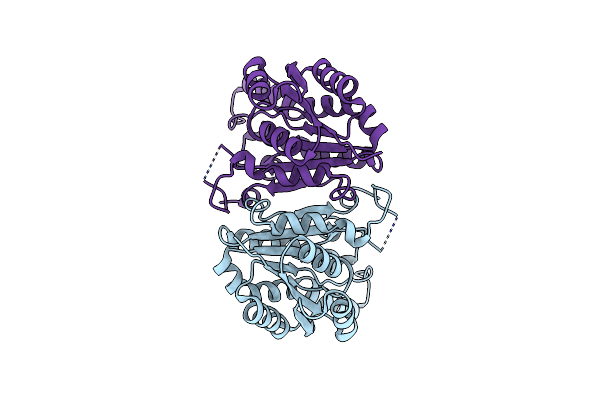 |
Crystal Structure Of The Cis-Dihydrodiol Naphthalene Dehydrogenase Nahb From Pseudomonas Sp. Mc1
Organism: Pseudomonas sp. mc1
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2017-08-09 Classification: OXIDOREDUCTASE |
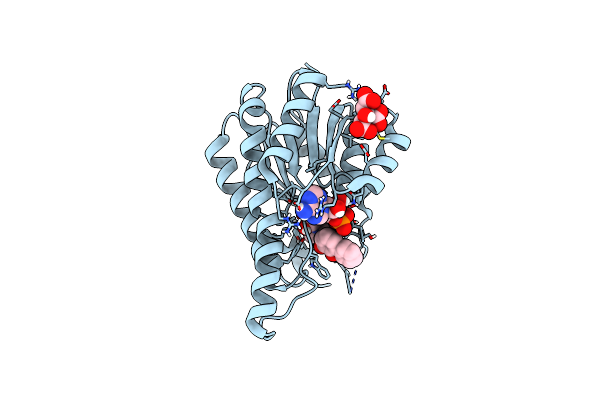 |
Crystal Structure Of The Cis-Dihydrodiol Naphthalene Dehydrogenase Nahb From Pseudomonas Sp. Mc1 In The Presence Of Nad+ And 2,3-Dihydroxybiphenyl
Organism: Pseudomonas sp. mc1
Method: X-RAY DIFFRACTION Resolution:2.32 Å Release Date: 2017-08-09 Classification: OXIDOREDUCTASE Ligands: NAD, CIT, BPY |
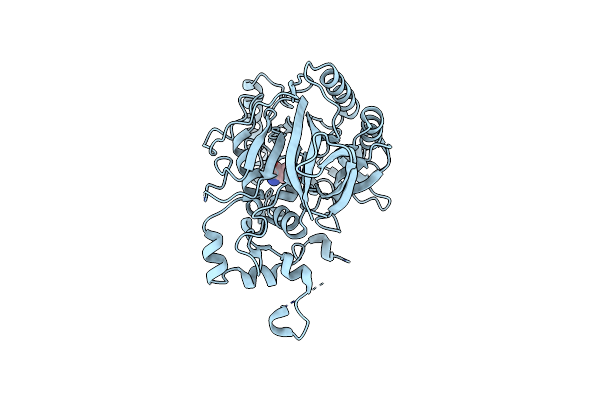 |
Organism: Saccharomyces cerevisiae (strain cen.pk113-7d)
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2017-04-19 Classification: HYDROLASE Ligands: GLN |
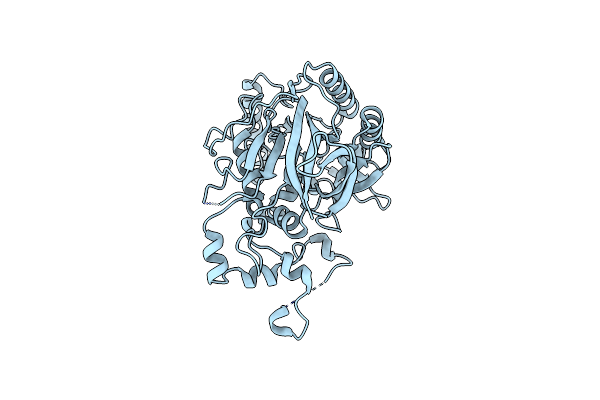 |
Organism: Saccharomyces cerevisiae (strain cen.pk113-7d)
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2017-04-05 Classification: HYDROLASE |
 |
Organism: Escherichia coli, Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2017-03-22 Classification: PROTEIN BINDING Ligands: ACT, GOL |
 |
Crystal Structures Of Aldehyde Deformylating Oxygenase From Limnothrix Sp. Knua012
Organism: Limnothrix sp. knua012
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2017-03-15 Classification: METAL BINDING PROTEIN Ligands: OCD |
 |
Organism: Saccharomyces cerevisiae (strain cen.pk113-7d), Saccharomyces cerevisiae cen.pk113-7d
Method: X-RAY DIFFRACTION Resolution:3.04 Å Release Date: 2017-01-11 Classification: HYDROLASE |
 |
Organism: Saccharomyces cerevisiae (strain cen.pk113-7d)
Method: X-RAY DIFFRACTION Resolution:2.32 Å Release Date: 2017-01-11 Classification: HYDROLASE |
 |
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2017-01-11 Classification: HYDROLASE |
 |
Organism: Saccharomyces cerevisiae (strain cen.pk113-7d)
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2017-01-11 Classification: HYDROLASE Ligands: GLN, VAL |
 |
Organism: Saccharomyces cerevisiae (strain cen.pk113-7d)
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2017-01-11 Classification: HYDROLASE Ligands: ASN, VAL |
 |
Organism: Saccharomyces cerevisiae (strain cen.pk113-7d)
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2017-01-11 Classification: HYDROLASE Ligands: ASN, GLY |
 |
Organism: Saccharomyces cerevisiae (strain cen.pk113-7d)
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2017-01-11 Classification: HYDROLASE Ligands: ASN, GLU |
 |
Structure And Catalytic Mechanism Of Monodehydroascorbate Reductase, Mdhar, From Oryza Sativa L. Japonica
Organism: Oryza sativa subsp. japonica
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2016-10-12 Classification: HYDROLASE Ligands: FAD |
 |
Structure And Catalytic Mechanism Of Monodehydroascorbate Reductase, Mdhar, From Oryza Sativa L. Japonica
Organism: Oryza sativa subsp. japonica
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2016-10-12 Classification: HYDROLASE Ligands: FAD, NAD |

