Search Count: 15
 |
Organism: Enhygromyxa salina
Method: X-RAY DIFFRACTION Release Date: 2024-11-27 Classification: OXIDOREDUCTASE Ligands: CD |
 |
Organism: Adeno-associated virus - 8
Method: ELECTRON MICROSCOPY Release Date: 2021-07-07 Classification: VIRUS Ligands: D5M |
 |
Organism: Bacillus cereus
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2017-09-06 Classification: OXIDOREDUCTASE |
 |
Organism: Bacillus cereus
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2017-09-06 Classification: OXIDOREDUCTASE Ligands: NAD, NA |
 |
Organism: Bacillus cereus
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2017-09-06 Classification: OXIDOREDUCTASE |
 |
Organism: Bacillus cereus
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2015-05-20 Classification: OXIDOREDUCTASE |
 |
Organism: Bacillus cereus
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2015-05-20 Classification: OXIDOREDUCTASE |
 |
Organism: Clostridium thermocellum
Method: X-RAY DIFFRACTION Resolution:2.07 Å Release Date: 2011-06-22 Classification: ISOMERASE Ligands: RB5 |
 |
Organism: Clostridium thermocellum
Method: X-RAY DIFFRACTION Resolution:2.07 Å Release Date: 2011-04-13 Classification: ISOMERASE Ligands: AOS |
 |
Organism: Escherichia coli o157:h7
Method: X-RAY DIFFRACTION Resolution:1.58 Å Release Date: 2010-07-21 Classification: ISOMERASE Ligands: GOL, ACT, MN, IPH |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.91 Å Release Date: 2010-07-21 Classification: ISOMERASE Ligands: MN, ACT, FRU, GOL |
 |
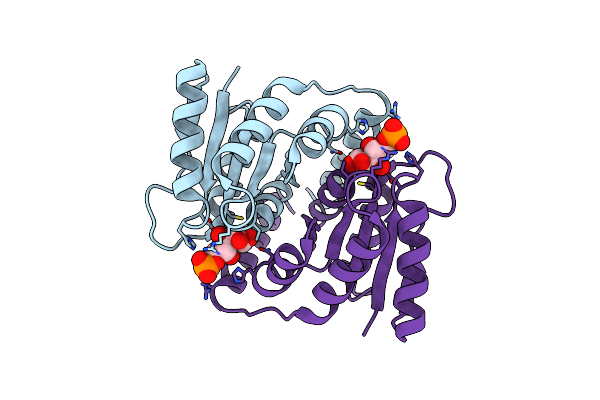
Structural Study Of Clostridium Thermocellum Ribose-5-Phosphate Isomerase B
Organism: Clostridium thermocellum
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2009-11-10 Classification: ISOMERASE Ligands: GOL |
 |
Structural Study Of Clostridium Thermocellum Ribose-5-Phosphate Isomerase B And Ribose-5-Phosphate
Organism: Clostridium thermocellum
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2009-11-10 Classification: ISOMERASE Ligands: R5P |
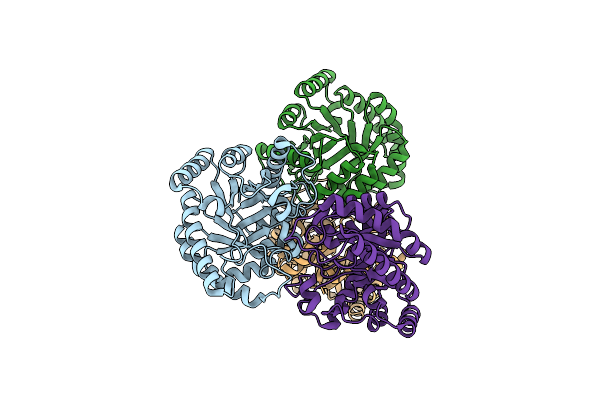 |
Crystal Structure Of D-Psicose 3-Epimerase (Dpease) In The Absence Of Substrate
Organism: Agrobacterium tumefaciens
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2006-08-29 Classification: ISOMERASE |
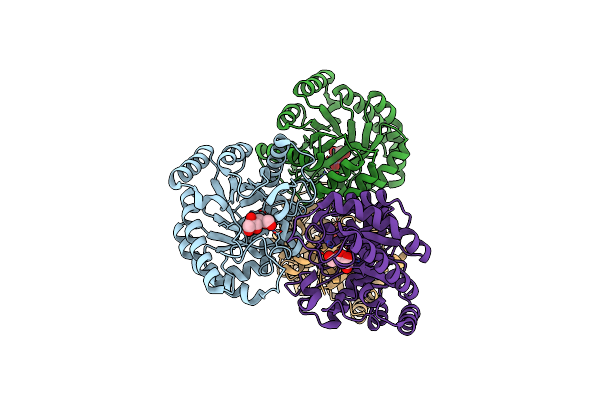 |
Crystal Structure Of D-Psicose 3-Epimerase (Dpease) In The Presence Of D-Fructose
Organism: Agrobacterium tumefaciens
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2006-08-29 Classification: ISOMERASE Ligands: FUD, MN |

