Search Count: 29
 |
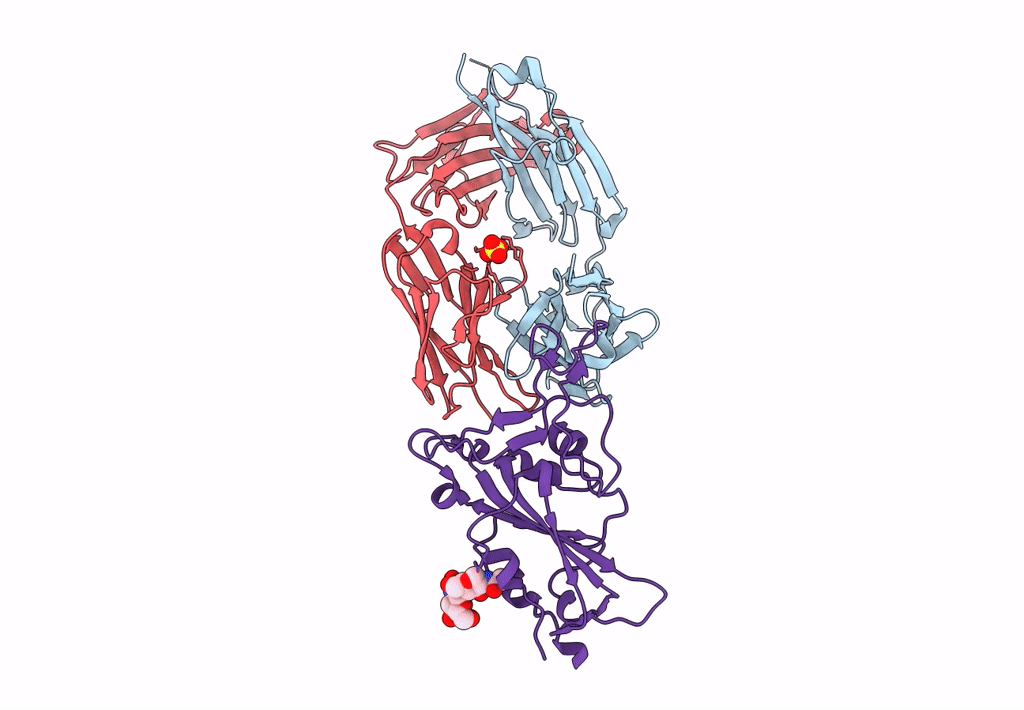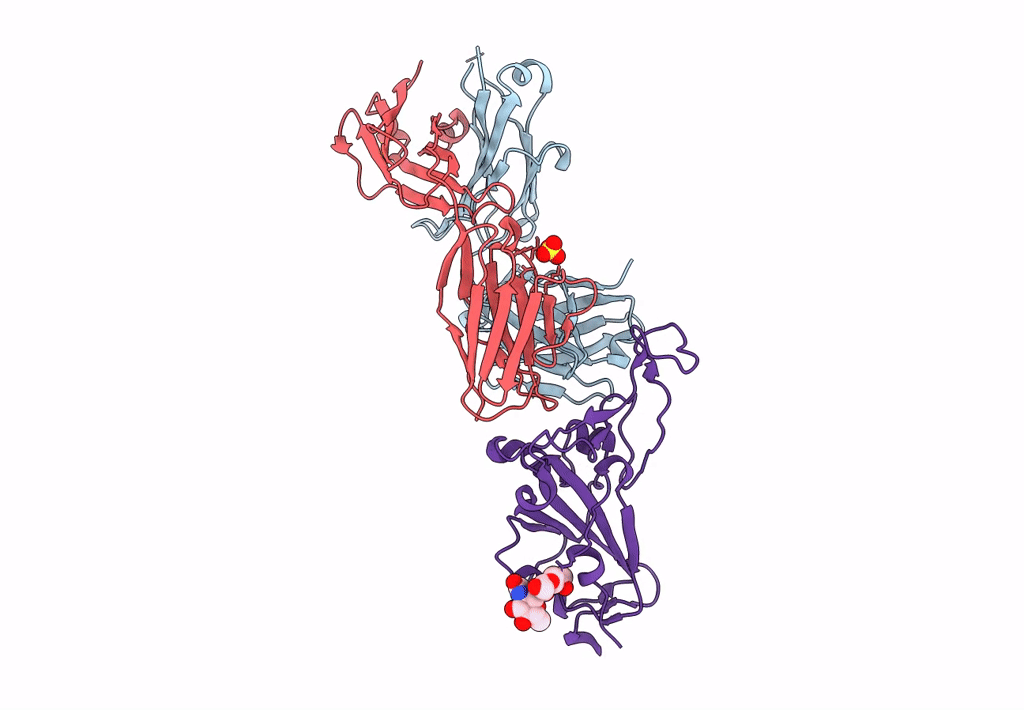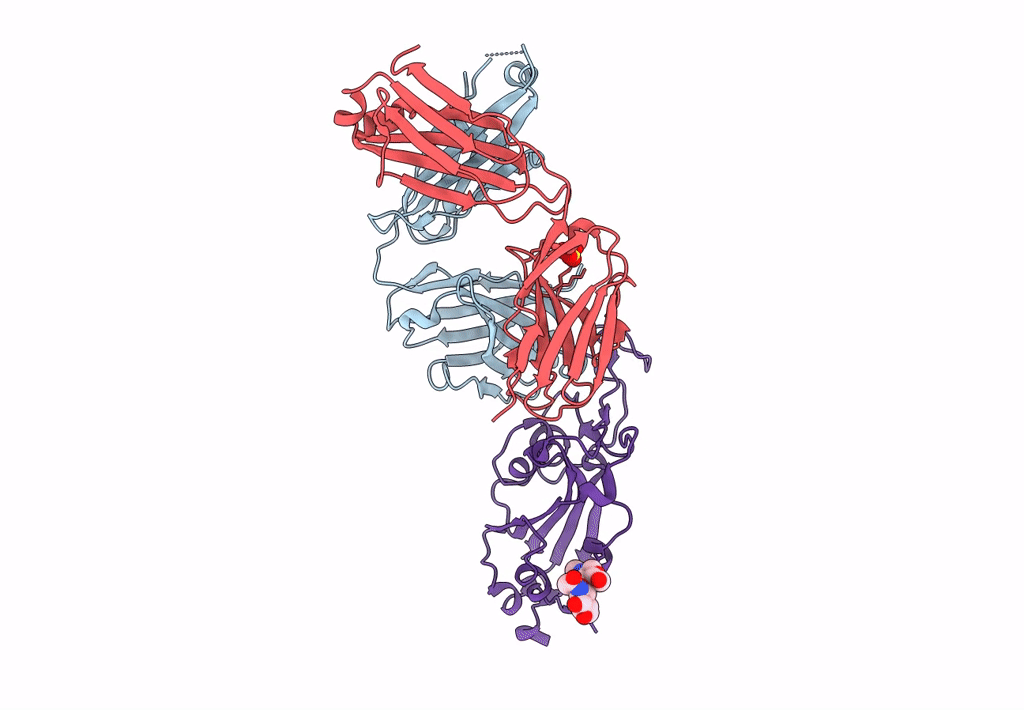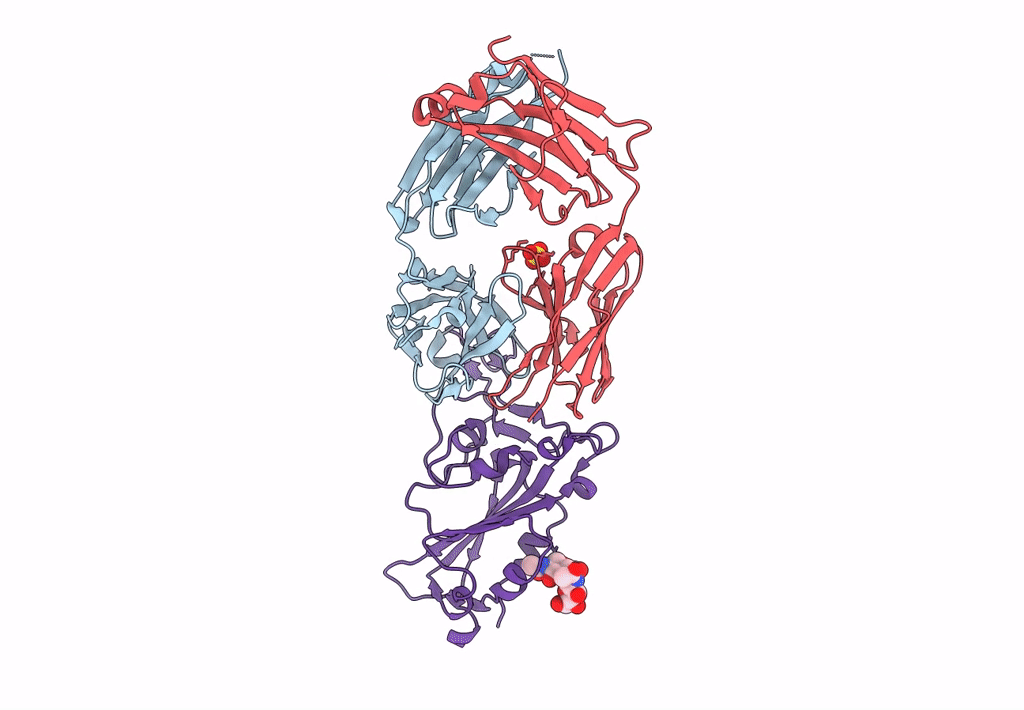
Crystal Structure Of Sars-Cov-2 Spike Receptor-Binding Domain (Rbd) In Complex With Ncv2Sg53 Fab
Organism: Severe acute respiratory syndrome coronavirus 2, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.35 Å Release Date: 2023-04-19 Classification: VIRAL PROTEIN/IMMUNE SYSTEM Ligands: SO4, NAG |
 |
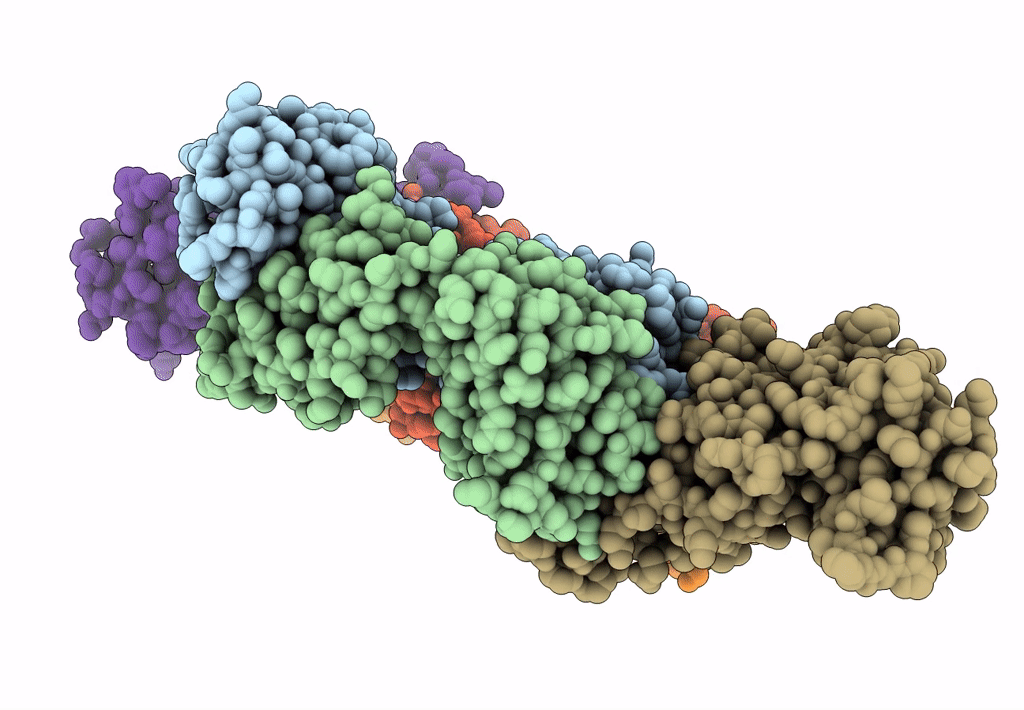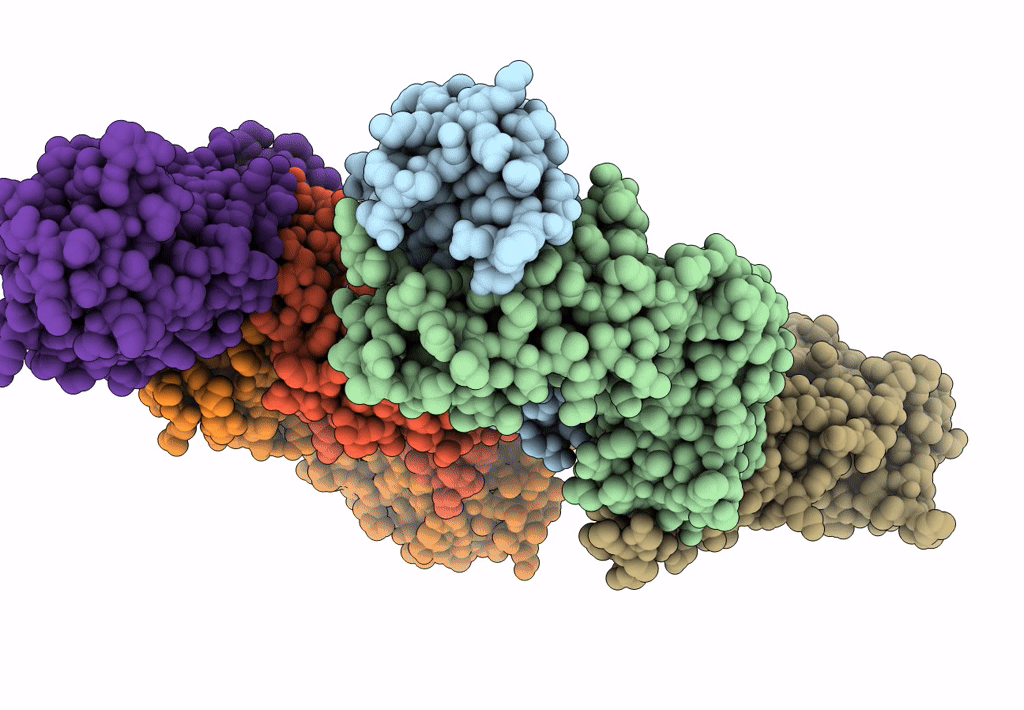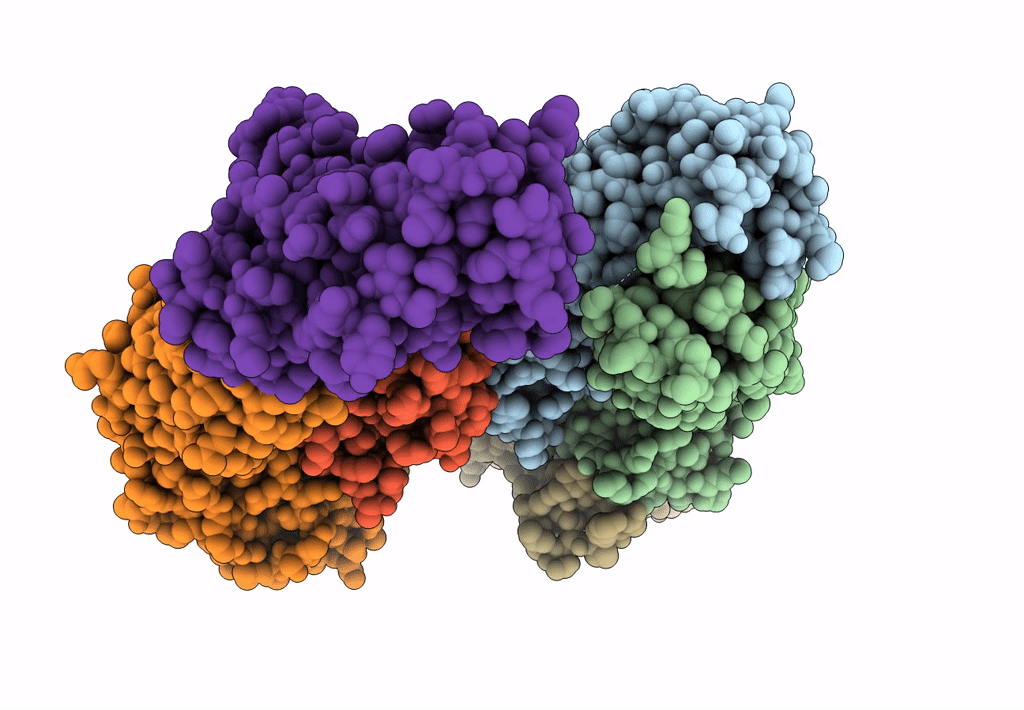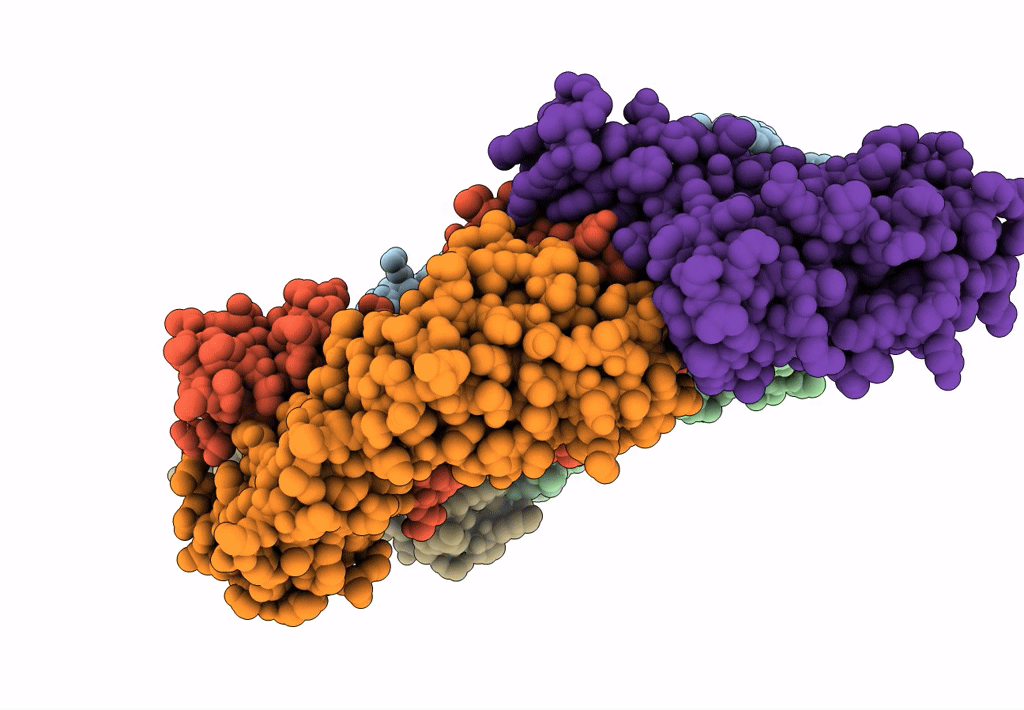
Crystal Structure Of Sars-Cov-2 Spike Receptor-Binding Domain (Rbd) In Complex With Ncv2Sg48 Fab
Organism: Severe acute respiratory syndrome coronavirus 2, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.18 Å Release Date: 2023-04-19 Classification: VIRAL PROTEIN/IMMUNE SYSTEM Ligands: SO4 |
 |
Crystal Structure Of Sars-Cov-2 Omicron Variant Spike Receptor-Binding Domain (Rbd) In Complex With Ncv2Sg48 Fab
Organism: Homo sapiens, Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:3.30 Å Release Date: 2023-04-19 Classification: IMMUNE SYSTEM/VIRAL PROTEIN Ligands: SO4 |
 |
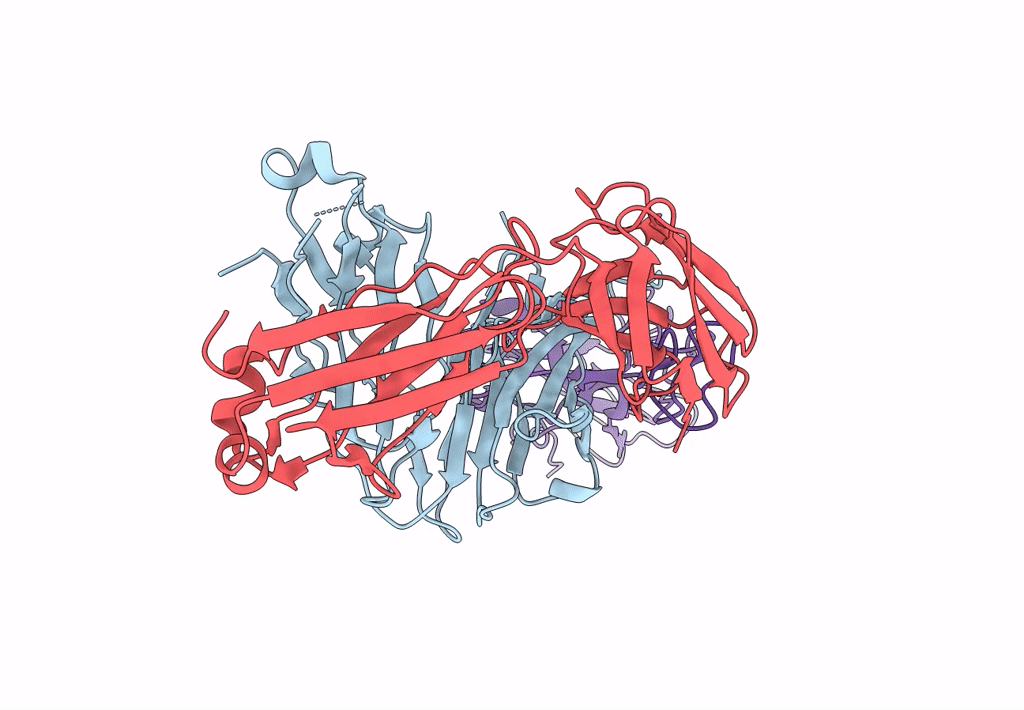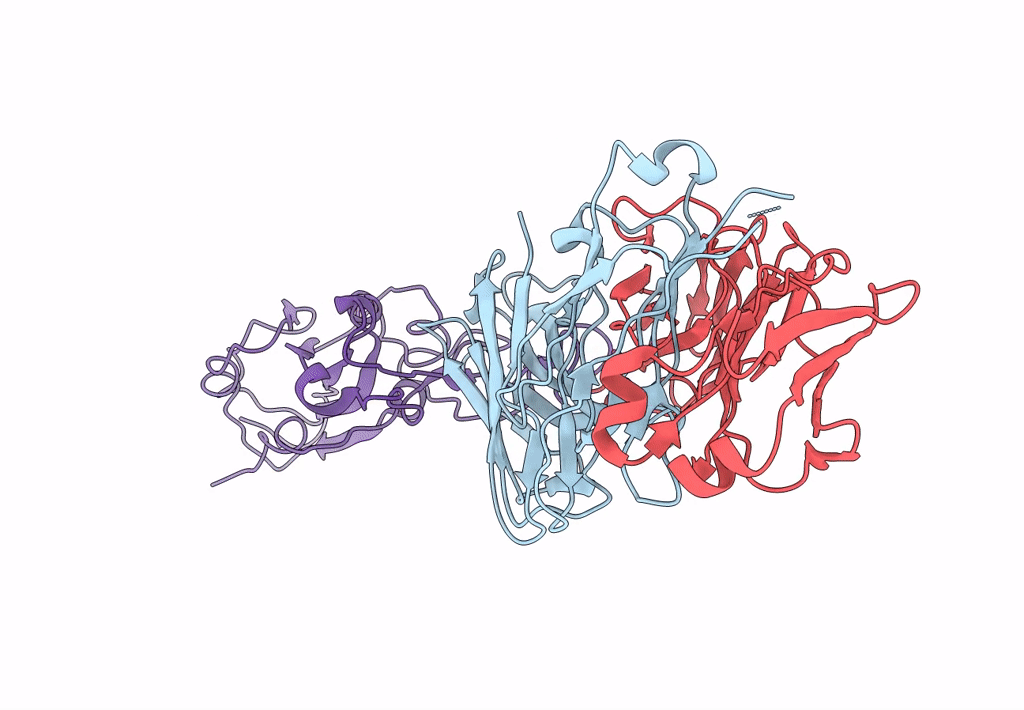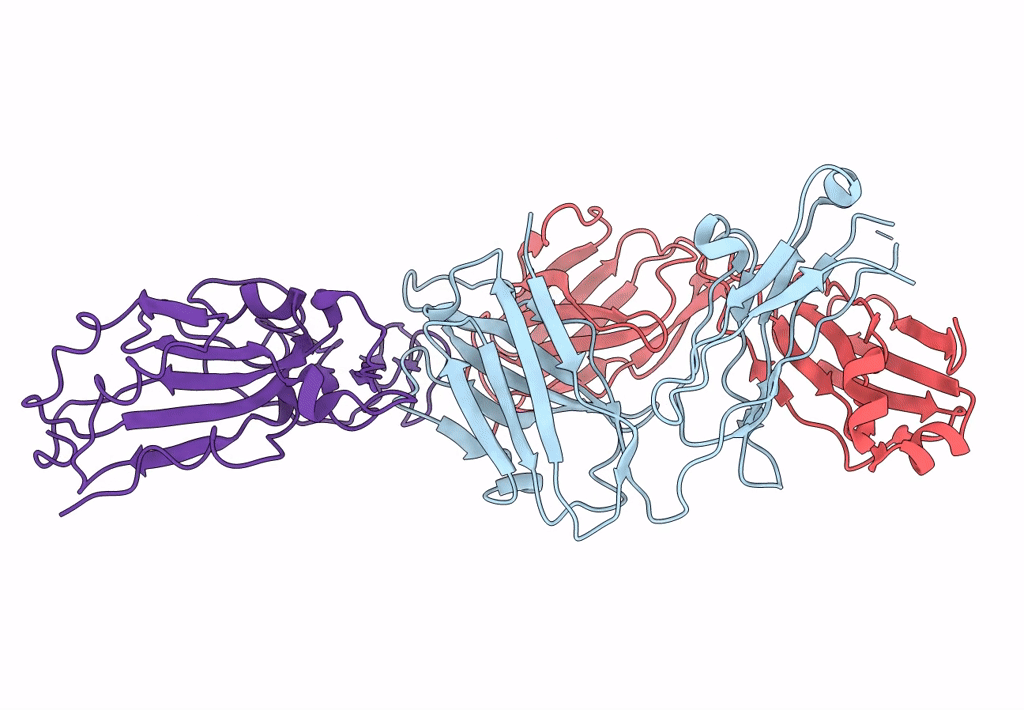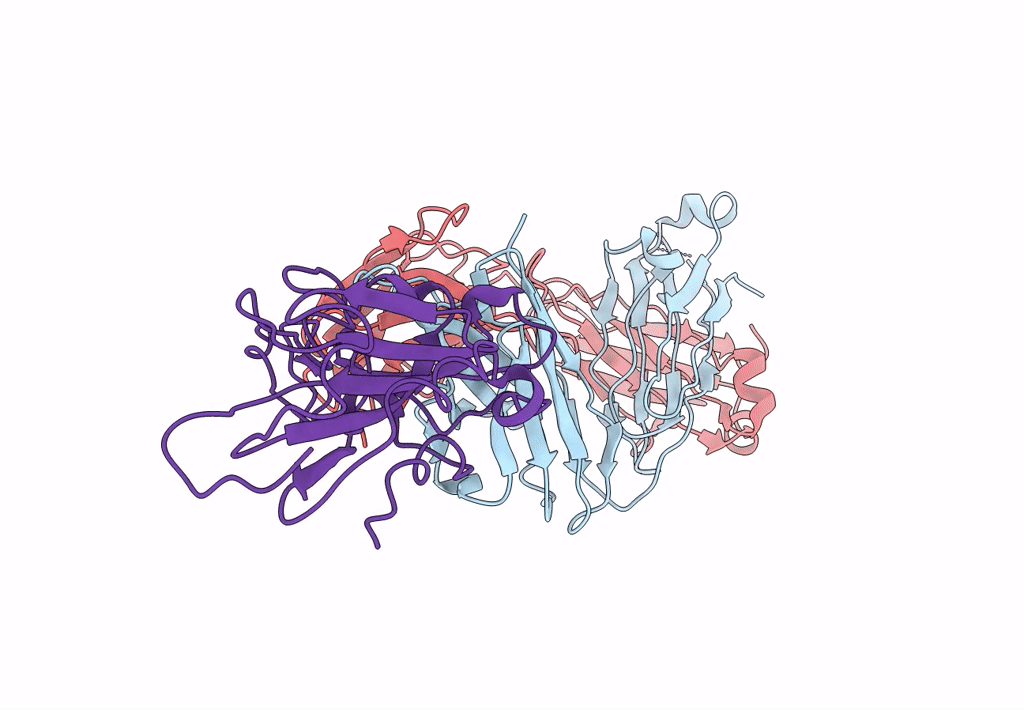
Crystal Structure Of Sars-Cov-2 Delta Variant Spike Receptor-Binding Domain (Rbd) In Complex With Ncv2Sg48 Fab
Organism: Severe acute respiratory syndrome coronavirus 2, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.38 Å Release Date: 2023-04-19 Classification: VIRAL PROTEIN/IMMUNE SYSTEM Ligands: NAG |
 |
Crystal Structure Of Sars-Cov-2 Delta Variant Spike Receptor-Binding Domain (Rbd) In Complex With Ncv2Sg53 Fab
Organism: Severe acute respiratory syndrome coronavirus 2, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.06 Å Release Date: 2023-04-19 Classification: VIRAL PROTEIN/IMMUNE SYSTEM |
 |
Structure Of Mcm2-7 Dh Complexed With Cdc7-Dbf4 In The Presence Of Atpgs, State Iii
Organism: Saccharomyces cerevisiae (strain atcc 204508 / s288c)
Method: ELECTRON MICROSCOPY Release Date: 2022-06-08 Classification: REPLICATION Ligands: AGS, MG, ZN, ADP |
 |
Structure Of Mcm2-7 Dh Complexed With Cdc7-Dbf4 In The Presence Of Adp:Bef3, State I
Organism: Saccharomyces cerevisiae (strain atcc 204508 / s288c)
Method: ELECTRON MICROSCOPY Release Date: 2022-06-08 Classification: REPLICATION Ligands: ADP, MG, ZN, BEF |
 |
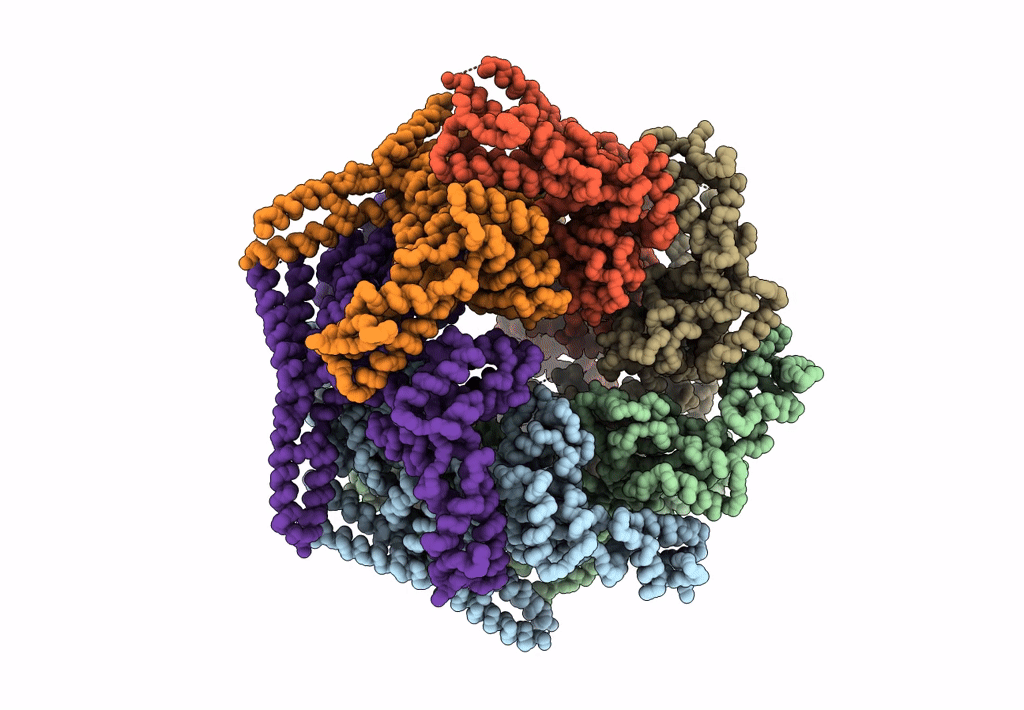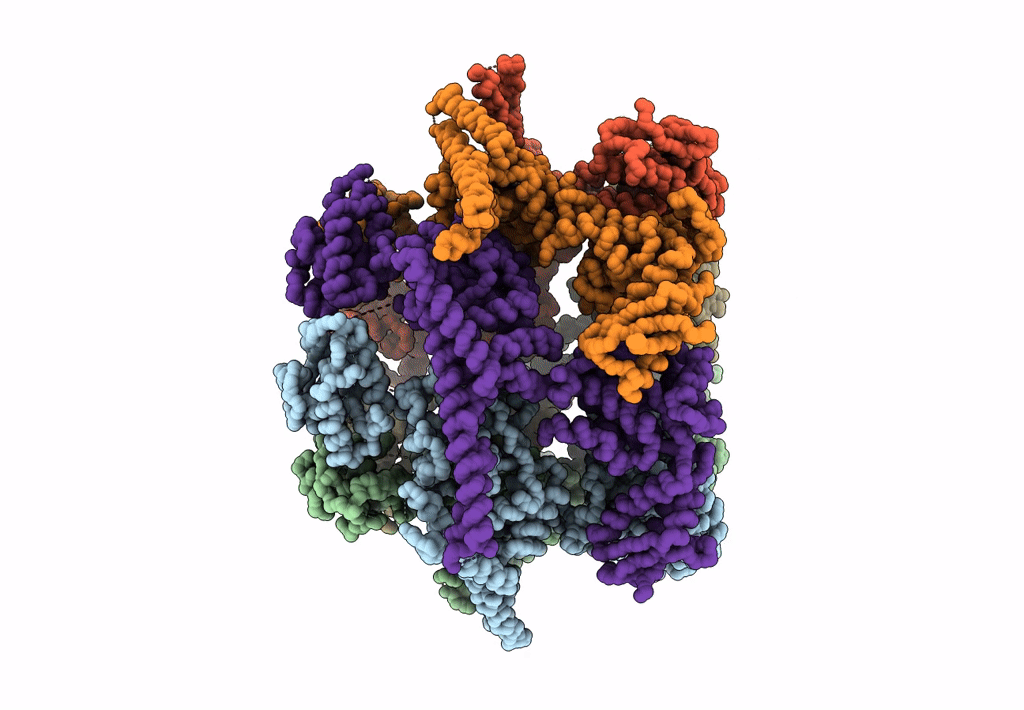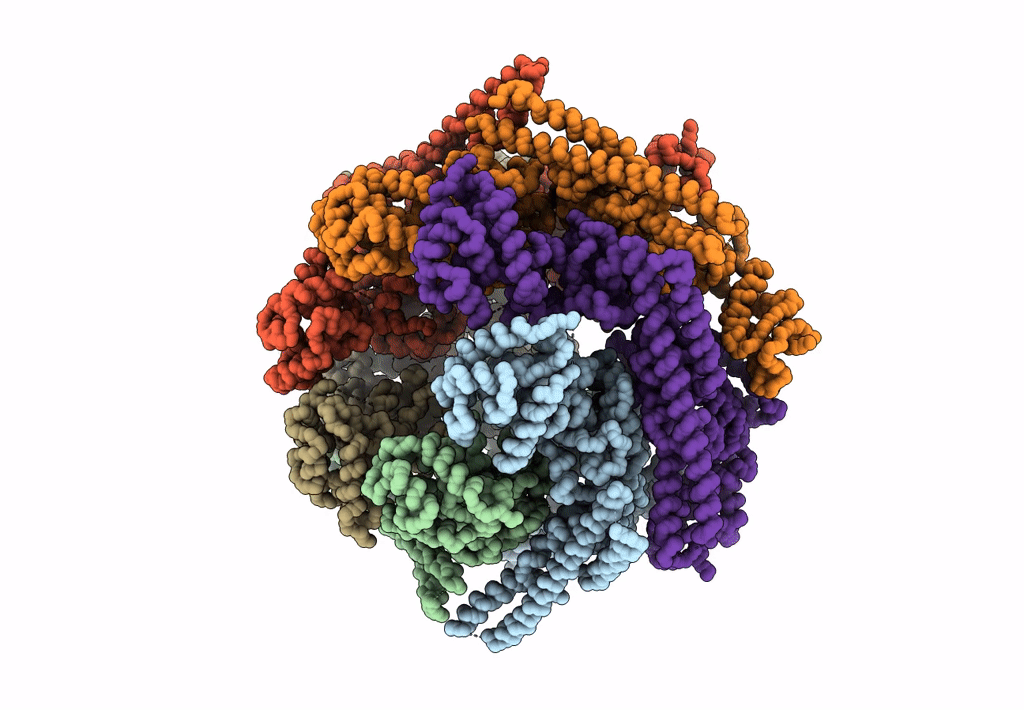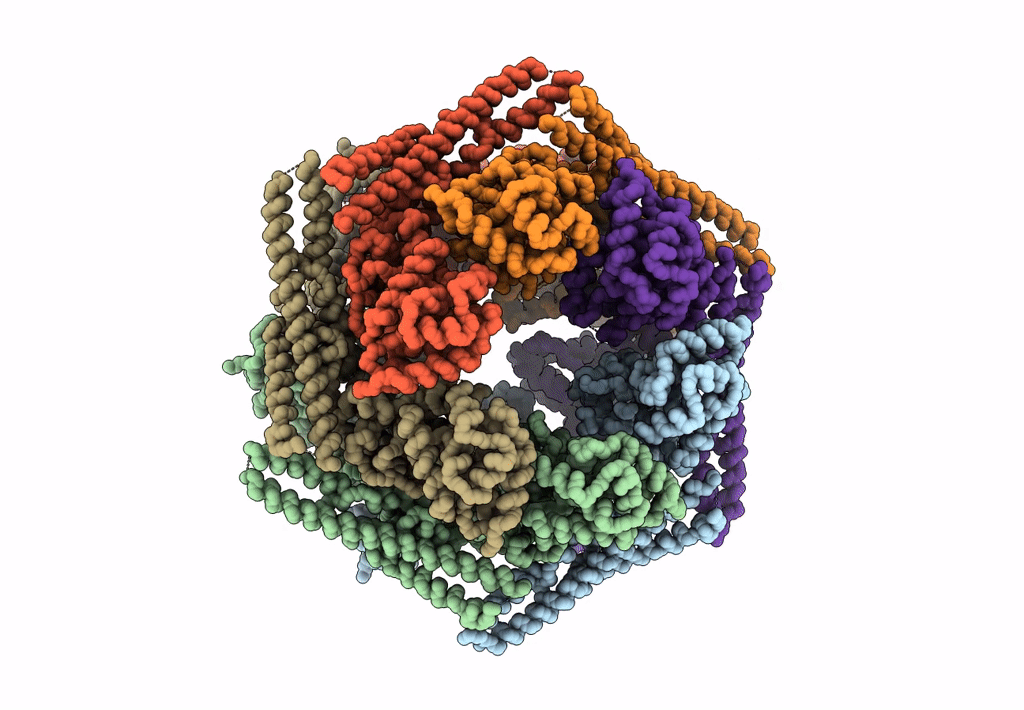
Structure Of The S. Cerevisiae Origin Recognition Complex Bound To The Replication Initiator Cdc6 And The Ars1 Origin Dna.
Organism: Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2021-06-02 Classification: REPLICATION Ligands: MG, AGS |
 |
Organism: Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2017-10-25 Classification: HYDROLASE/DNA Ligands: ADP |
 |
Negative-Stain Electron Microscopy Of E. Coli Clpb Mutant E432A (Bap Form Bound To Clpp)
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Resolution:18.00 Å Release Date: 2014-06-04 Classification: CHAPERONE |
 |
Negative-Stain Electron Microscopy Of E. Coli Clpb (Bap Form Bound To Clpp)
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Resolution:17.00 Å Release Date: 2014-06-04 Classification: CHAPERONE |
 |
Negative-Stain Electron Microscopy Of E. Coli Clpb Of Y503D Hyperactive Mutant (Bap Form Bound To Clpp)
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Resolution:20.00 Å Release Date: 2014-06-04 Classification: CHAPERONE |
 |
Crystal Structure Of Bacillus Subtilis Ywfe, An L-Amino Acid Ligase, With Bound Adp-Mg-Pi
Organism: Bacillus subtilis
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2014-05-14 Classification: LIGASE Ligands: ADP, MG, PO4 |
 |
Crystal Structure Of Bacillus Subtilis Ywfe, An L-Amino Acid Ligase, With Bound Adp-Mg-Ala
Organism: Bacillus subtilis
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2014-05-14 Classification: LIGASE Ligands: ADP, MG, ALA |
 |
Crystal Structure Of Trp332Ala Mutant Ywfe, An L-Amino Acid Ligase, With Bound Adp-Mg-Ala
Organism: Bacillus subtilis
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2014-05-14 Classification: LIGASE Ligands: ADP, MG, ALA |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:3.50 Å Release Date: 2014-05-14 Classification: CHAPERONE Ligands: ADP |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.35 Å Release Date: 2008-08-05 Classification: PROTEIN TRANSPORT |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2008-08-05 Classification: PROTEIN TRANSPORT |
 |
Human Telomeric Dna Mixed-Parallel/Antiparallel Quadruplex Under Physiological Ionic Conditions Stabilized By Proper Incorporation Of 8-Bromoguanosines
|
 |
Three-Dimensional Structure Of The Carbon Monoxide Complex Of [Nife]Hydrogenase From Desulufovibrio Vulgaris Miyazaki F
Organism: Desulfovibrio vulgaris str. 'miyazaki f'
Method: X-RAY DIFFRACTION Resolution:1.35 Å Release Date: 2003-04-29 Classification: OXIDOREDUCTASE Ligands: SF4, F3S, MPD, MG, FNE, CMO |

