Search Count: 33
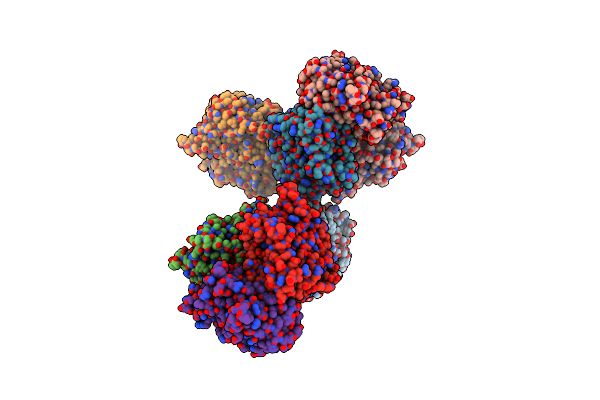 |
Crystal Structure Of A Dodecameric Multicopper Oxidase From M. Hydrothermalis In A Cubic Lattice
Organism: Marinithermus hydrothermalis
Method: X-RAY DIFFRACTION Resolution:1.92 Å Release Date: 2021-10-20 Classification: OXIDOREDUCTASE Ligands: CU, MG |
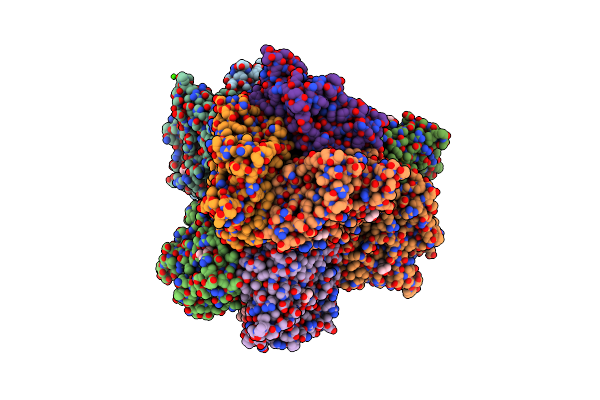 |
Crystal Structure Of A Dodecameric Multicopper Oxidase From M. Hydrothermalis In An Orthorhombic Lattice
Organism: Marinithermus hydrothermalis
Method: X-RAY DIFFRACTION Resolution:2.36 Å Release Date: 2021-10-20 Classification: OXIDOREDUCTASE Ligands: MPD, CU, CA |
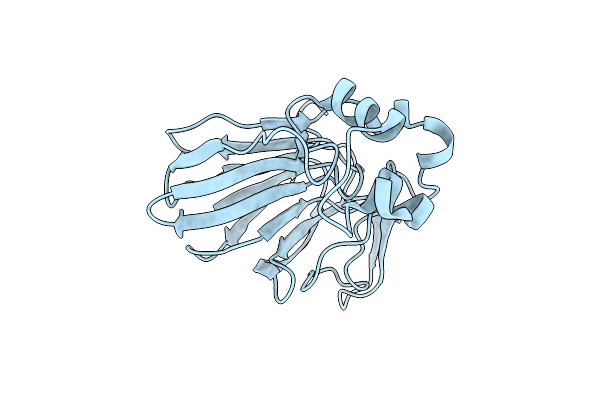 |
Organism: Thaumatococcus daniellii
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2018-05-30 Classification: PLANT PROTEIN |
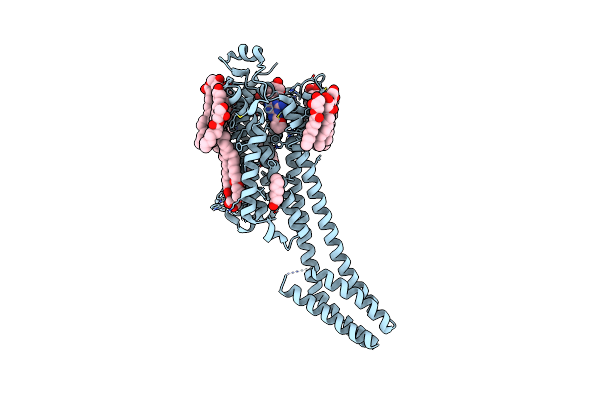 |
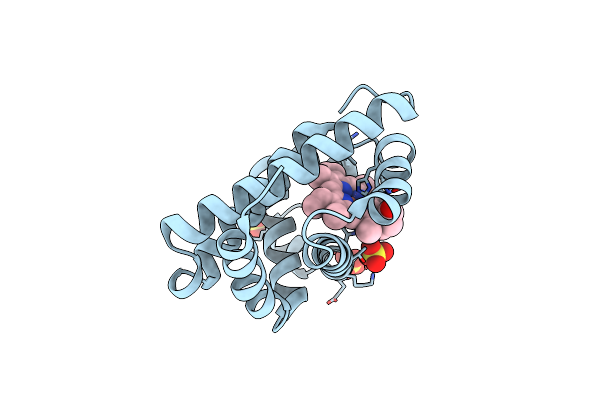
2.35-Angstrom In Situ Mylar Structure Of Human A2A Adenosine Receptor At 100 K
Organism: Homo sapiens, Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.35 Å Release Date: 2017-12-13 Classification: SIGNALING PROTEIN Ligands: ZMA, NA, CLR, OLC, OLA, OLB |
 |
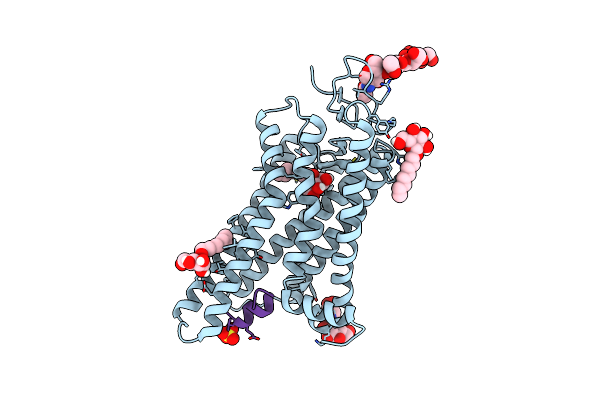
2.0-Angstrom In Situ Mylar Structure Of Sperm Whale Myoglobin (Swmb) At 293 K
Organism: Physeter catodon
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2017-12-13 Classification: OXYGEN TRANSPORT Ligands: HEM, CL, SO4 |
 |
Organism: Bos taurus
Method: X-RAY DIFFRACTION Resolution:3.20 Å Release Date: 2017-12-13 Classification: SIGNALING PROTEIN Ligands: BOG, SO4 |
 |
Organism: Engyodontium album
Method: X-RAY DIFFRACTION Resolution:2.21 Å Release Date: 2017-11-15 Classification: HYDROLASE Ligands: CA, CL, NHE, EPE |
 |
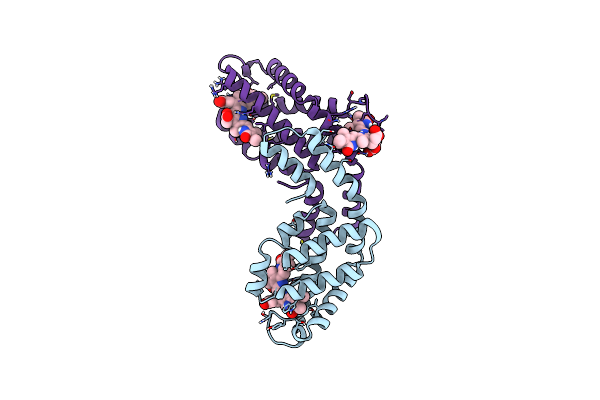
Single-Shot Pink Beam Serial Crystallography: Phycocyanin (Five Chips Merged)
Organism: Thermosynechococcus elongatus (strain bp-1), Synechococcus elongatus
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2017-11-15 Classification: PHOTOSYNTHESIS Ligands: CYC |
 |
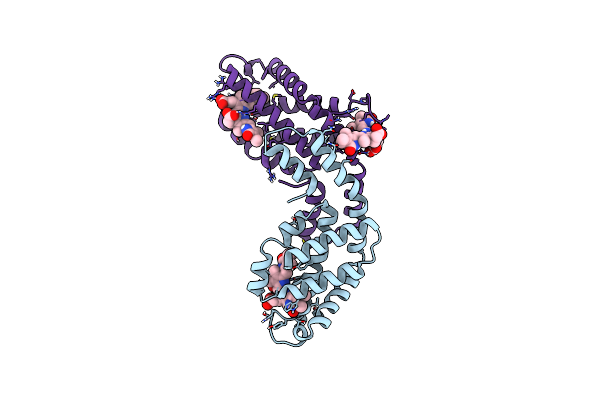
Organism: Thermosynechococcus elongatus (strain bp-1)
Method: X-RAY DIFFRACTION Resolution:2.11 Å Release Date: 2017-11-15 Classification: PHOTOSYNTHESIS Ligands: CYC |
 |
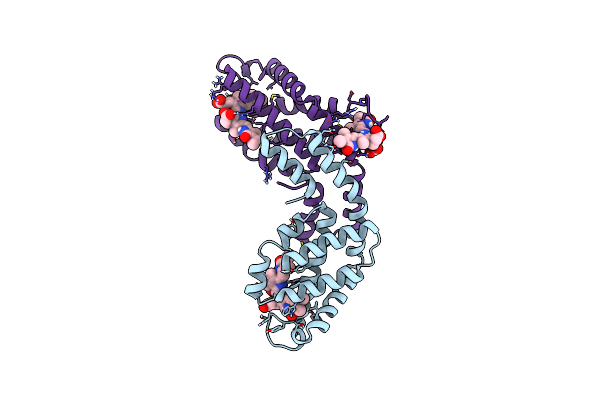
Organism: Thermosynechococcus elongatus (strain bp-1)
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2017-11-15 Classification: PHOTOSYNTHESIS Ligands: CYC |
 |
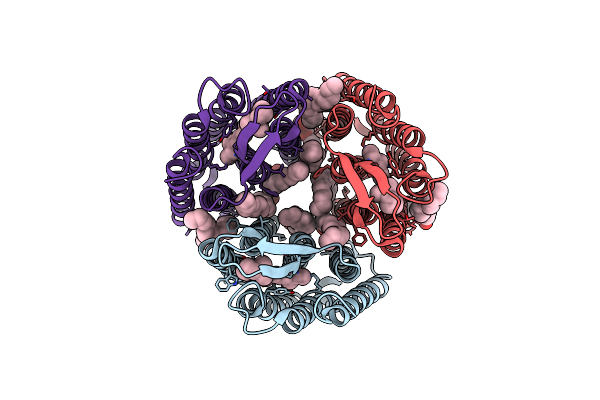
Single-Shot Pink Beam Serial Crystallography: Phycocyanin (One Chip, Chip_1)
Organism: Thermosynechococcus elongatus (strain bp-1)
Method: X-RAY DIFFRACTION Resolution:2.46 Å Release Date: 2017-11-15 Classification: PHOTOSYNTHESIS Ligands: CYC |
 |
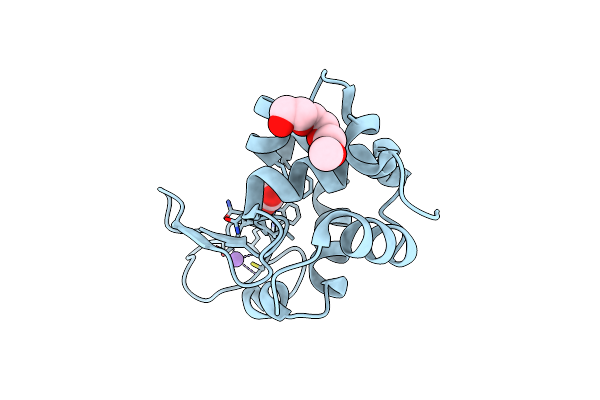
2.1-Angstrom In Situ Mylar Structure Of Bacteriorhodopsin From Haloquadratum Walsbyi (Hwbr) At 100 K
Organism: Haloquadratum walsbyi (strain dsm 16790 / hbsq001)
Method: X-RAY DIFFRACTION Resolution:2.13 Å Release Date: 2017-02-15 Classification: MEMBRANE PROTEIN Ligands: OLC, OLB, RET |
 |
1.7-Angstrom In Situ Mylar Structure Of Hen Egg-White Lysozyme (Hewl) At 100 K
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2017-02-15 Classification: HYDROLASE |
 |
2.0-Angstrom In Situ Mylar Structure Of Hen Egg-White Lysozyme (Hewl) At 293 K
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2017-02-15 Classification: HYDROLASE |
 |
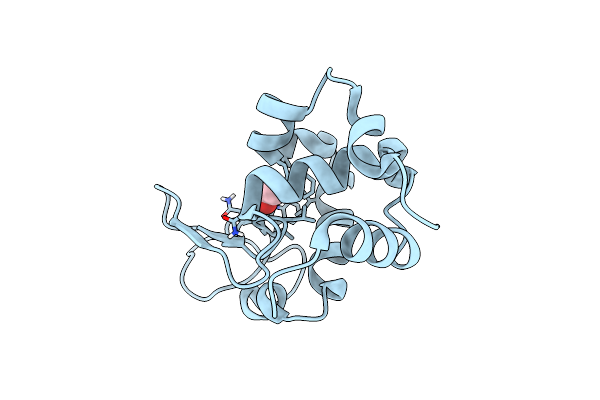
1.7-Angstrom In Situ Mylar Structure Of Sperm Whale Myoglobin (Swmb-Co) At 100 K
Organism: Physeter catodon
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2017-02-15 Classification: OXYGEN TRANSPORT Ligands: HEM, CMO, SO4, CL |
 |
Crystal Structure Of Hen Egg White Lysozyme Using Serial X-Ray Diffraction Data Collection
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2015-10-14 Classification: HYDROLASE Ligands: ACT |
 |
Organism: West nile virus
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2014-10-15 Classification: VIRAL PROTEIN Ligands: NAG, SO4, OXN |
 |
Organism: Dengue virus 2
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2014-02-19 Classification: VIRAL PROTEIN Ligands: NAG |
 |
Organism: West nile virus
Method: X-RAY DIFFRACTION Resolution:2.75 Å Release Date: 2014-02-19 Classification: VIRAL PROTEIN Ligands: NAG, SO4 |
 |
Organism: West nile virus
Method: X-RAY DIFFRACTION Resolution:2.59 Å Release Date: 2014-02-19 Classification: VIRAL PROTEIN Ligands: NAG, OXN, SO4 |

