Search Count: 54
 |
Non-Crystalline Proximal Tubulopathy And Crystalline Cast Nephropathy-Causing Bence-Jones Protein Pt-Cn: An Entire Immunoglobulin Kappa Light Chain Dimer
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2025-01-22 Classification: IMMUNE SYSTEM Ligands: PO4, ETX, PGE, GOL, EDO, PEG |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.30 Å Release Date: 2024-03-13 Classification: CYTOKINE Ligands: SO4, EDO, IPA |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION, NEUTRON DIFFRACTION Resolution:1.60 Å, 2.00 Å Release Date: 2023-05-31 Classification: CYTOKINE |
 |
High Resolution Crystal Structure Of Human Macrophage Migration Inhibitory Factor In Complex With Methotrexate
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.28 Å Release Date: 2023-05-24 Classification: CYTOKINE/INHIBITOR Ligands: SO4, EDO, IPA, MT1 |
 |
Crystalline Cast Nephropathy-Causing Bence-Jones Protein Ak: An Entire Immunoglobulin Lambda Light Chain Dimer
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.77 Å Release Date: 2020-09-23 Classification: IMMUNE SYSTEM |
 |
Crystal Structure Of The B-Type Halohydrin Hydrogen-Halide-Lyase Mutant F71W/Q125T/D199H From Corynebacterium Sp. N-1074
Organism: Corynebacterium sp.
Method: X-RAY DIFFRACTION Resolution:1.72 Å Release Date: 2016-08-03 Classification: LYASE Ligands: CL |
 |
Organism: Picrophilus torridus (strain atcc 700027 / dsm 9790 / jcm 10055 / nbrc 100828)
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2016-06-22 Classification: ISOMERASE Ligands: TLA |
 |
Crystal Structure Of Nitrile Hydratase Mutant Br56K Complexed With Trimethylacetonitrile, Photo-Activated For 25 Min
Organism: Rhodococcus erythropolis
Method: X-RAY DIFFRACTION Resolution:1.18 Å Release Date: 2016-01-27 Classification: LYASE Ligands: FE, TAN, MG, CL |
 |
Crystal Structure Of Nitrile Hydratase Mutant Br56K Complexed With Trimethylacetonitrile, Photo-Activated For 120 Min
Organism: Rhodococcus erythropolis
Method: X-RAY DIFFRACTION Resolution:1.24 Å Release Date: 2016-01-27 Classification: LYASE Ligands: FE, MG, TAN |
 |
Crystal Structure Of Nitrile Hydratase Mutant Br56K Complexed With Trimethylacetonitrile, Photo-Activated For 700 Min
Organism: Rhodococcus erythropolis
Method: X-RAY DIFFRACTION Resolution:1.20 Å Release Date: 2016-01-27 Classification: LYASE Ligands: FE, TAN, CL, MG |
 |
Crystal Structure Of Nitrile Hydratase Mutant Br56K Complexed With Trimethylacetonitrile, Photo-Activated For 5 Min
Organism: Rhodococcus erythropolis
Method: X-RAY DIFFRACTION Resolution:1.34 Å Release Date: 2016-01-27 Classification: LYASE Ligands: FE, MG, CL, TAN |
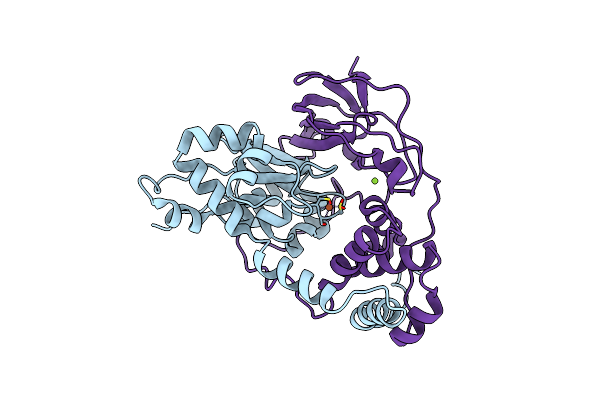 |
Organism: Rhodococcus erythropolis
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2015-12-23 Classification: LYASE Ligands: FE, MG, CL |
 |
Organism: Corynebacterium sp.
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2015-11-04 Classification: LYASE Ligands: CL |
 |
Organism: Corynebacterium sp.
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2015-11-04 Classification: LYASE Ligands: CL, MES, MG |
 |
Organism: Corynebacterium sp.
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2015-11-04 Classification: LYASE Ligands: 4SD, SO4, MG |
 |
Crystal Structure Of Nitrile Hydratase Mutant Br56K Complexed With Trimethylacetonitrile, Photo-Activated For 50 Min
Organism: Rhodococcus erythropolis
Method: X-RAY DIFFRACTION Resolution:1.18 Å Release Date: 2015-06-17 Classification: LYASE Ligands: FE, MG, TAN |
 |
Crystal Structure Of Nitrile Hydratase Mutant Br56K Complexed With Trimethylacetonitrile, Before Photo-Activation
Organism: Rhodococcus erythropolis
Method: X-RAY DIFFRACTION Resolution:1.57 Å Release Date: 2015-06-17 Classification: LYASE Ligands: FE, NO, MG, CL, TAN |
 |
Organism: Thiobacillus thioparus
Method: X-RAY DIFFRACTION Resolution:1.72 Å Release Date: 2013-11-13 Classification: HYDROLASE Ligands: 3CO, TLA |
 |
Crystal Structure Of Aw116R Mutant Of Nitrile Hydratase From Pseudonocardia Thermophilla
Organism: Pseudonocardia thermophila
Method: X-RAY DIFFRACTION Resolution:1.63 Å Release Date: 2013-11-13 Classification: LYASE Ligands: CO |
 |
Crystal Structutre Of Thiobacillus Thioparus Thi115 Carbonyl Sulfide Hydrolase
Organism: Thiobacillus thioparus
Method: X-RAY DIFFRACTION Resolution:1.20 Å Release Date: 2013-02-27 Classification: HYDROLASE Ligands: ZN, NA |

