Search Count: 212
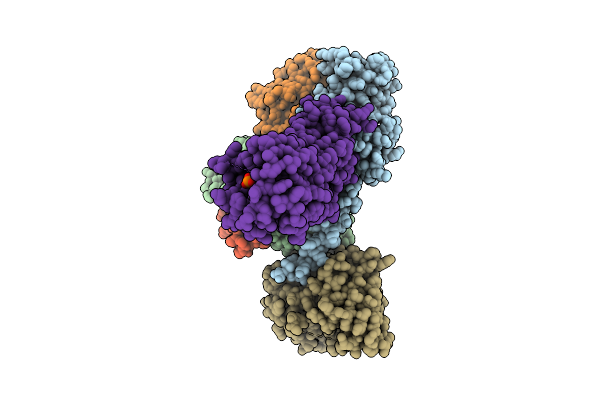 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Resolution:3.73 Å Release Date: 2025-11-26 Classification: MEMBRANE PROTEIN Ligands: A1L69 |
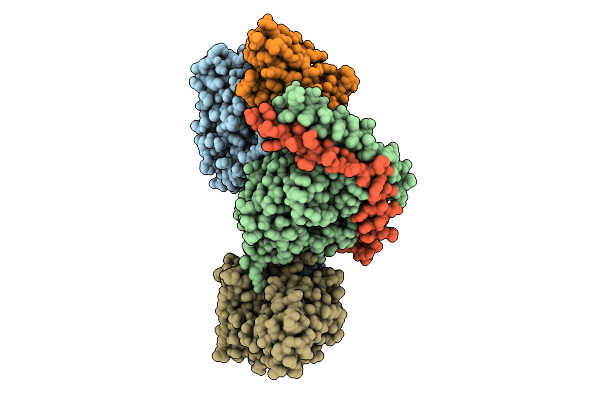 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Resolution:3.25 Å Release Date: 2025-11-26 Classification: MEMBRANE PROTEIN Ligands: S1P |
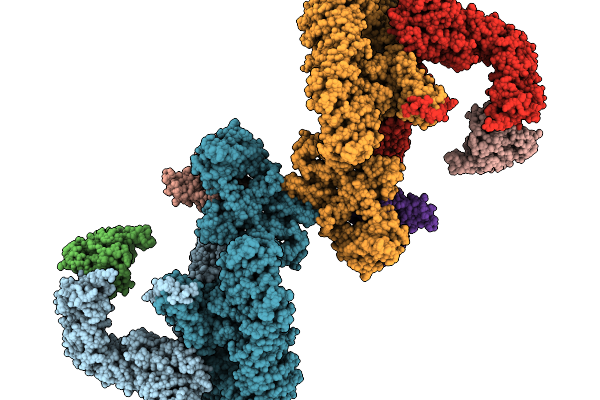 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Resolution:6.98 Å Release Date: 2025-11-19 Classification: SIGNALING PROTEIN |
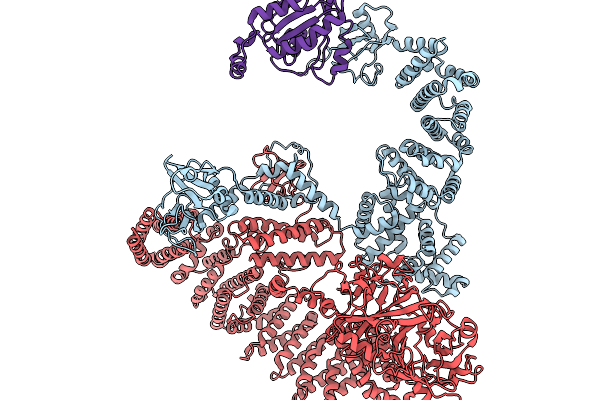 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Resolution:7.52 Å Release Date: 2025-11-19 Classification: SIGNALING PROTEIN |
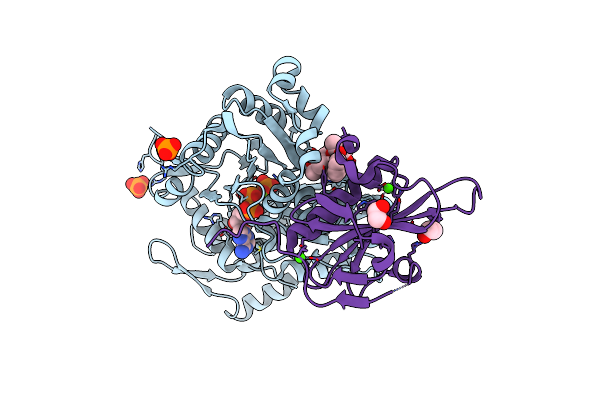 |
Crystal Structure Of Cytochalasin D Bound To A Filamentous Conformation Actin
Organism: Physarum polycephalum, Gallus gallus
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2025-07-02 Classification: CYTOSOLIC PROTEIN Ligands: ADP, PO4, MG, CY9, CA, EDO |
 |
Organism: Clostridium perfringens
Method: ELECTRON MICROSCOPY Release Date: 2025-05-14 Classification: TOXIN Ligands: CA |
 |
Organism: Clostridium perfringens
Method: ELECTRON MICROSCOPY Release Date: 2025-05-14 Classification: TOXIN Ligands: CA |
 |
Organism: Clostridium perfringens
Method: ELECTRON MICROSCOPY Release Date: 2025-05-14 Classification: TOXIN Ligands: CA |
 |
Organism: Clostridium perfringens
Method: ELECTRON MICROSCOPY Release Date: 2025-05-14 Classification: TOXIN Ligands: CA |
 |
Organism: Clostridium perfringens
Method: ELECTRON MICROSCOPY Release Date: 2025-05-14 Classification: TOXIN Ligands: CA |
 |
Organism: Clostridium perfringens
Method: ELECTRON MICROSCOPY Release Date: 2025-05-14 Classification: TOXIN Ligands: CA |
 |
Organism: Clostridium perfringens
Method: ELECTRON MICROSCOPY Release Date: 2025-05-14 Classification: TOXIN Ligands: CA |
 |
Organism: Clostridium perfringens
Method: ELECTRON MICROSCOPY Release Date: 2025-05-14 Classification: TOXIN Ligands: CA |
 |
Organism: Clostridium perfringens
Method: ELECTRON MICROSCOPY Release Date: 2025-05-14 Classification: TOXIN Ligands: CA |
 |
Organism: Zaire ebolavirus, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-01-29 Classification: VIRAL PROTEIN/RNA |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2024-11-06 Classification: IMMUNE SYSTEM |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-10-09 Classification: STRUCTURAL PROTEIN |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-10-09 Classification: STRUCTURAL PROTEIN |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-10-09 Classification: STRUCTURAL PROTEIN |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-10-09 Classification: STRUCTURAL PROTEIN |

