Search Count: 113
 |
Organism: Caenorhabditis elegans
Method: X-RAY DIFFRACTION Release Date: 2025-12-10 Classification: PROTEIN BINDING |
 |
Organism: Measles morbillivirus
Method: SOLUTION NMR Release Date: 2025-09-03 Classification: VIRUS Ligands: ZN |
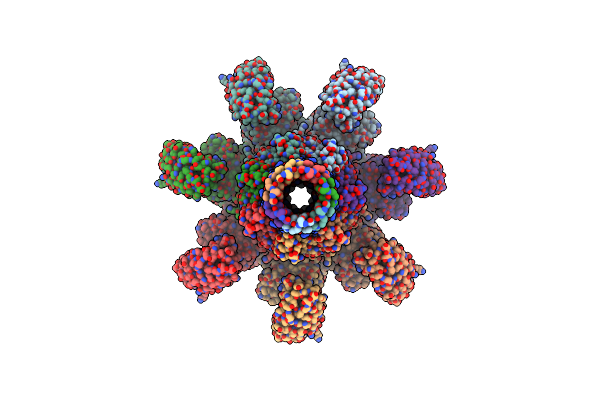 |
Organism: Clostridium perfringens
Method: ELECTRON MICROSCOPY Release Date: 2025-03-26 Classification: TOXIN Ligands: CA |
 |
Cryo-Em Structure Of Tetr Regulator Mce3R From Mycobacterium Tuberculosis Bound To A Dna Oligonucleotide
Organism: Mycobacterium tuberculosis h37rv
Method: ELECTRON MICROSCOPY Release Date: 2024-11-13 Classification: DNA BINDING PROTEIN/DNA |
 |
Organism: Klebsiella pneumoniae, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2023-08-02 Classification: CELL CYCLE Ligands: GDP |
 |
Organism: Klebsiella pneumoniae
Method: ELECTRON MICROSCOPY Release Date: 2023-08-02 Classification: CELL CYCLE Ligands: G2P, K |
 |
Organism: Klebsiella pneumoniae, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2023-07-19 Classification: CELL CYCLE Ligands: GDP |
 |
Organism: Klebsiella pneumoniae, Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2023-07-19 Classification: CELL CYCLE Ligands: GDP |
 |
Organism: Escherichia coli, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.84 Å Release Date: 2023-07-19 Classification: CELL CYCLE Ligands: GDP |
 |
Organism: Escherichia coli, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2023-07-19 Classification: CELL CYCLE Ligands: GDP |
 |
Organism: Schizosaccharomyces pombe (strain 972 / atcc 24843)
Method: X-RAY DIFFRACTION Resolution:1.58 Å Release Date: 2023-05-17 Classification: LIPID BINDING PROTEIN Ligands: 3PE |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2022-12-07 Classification: LIGASE |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.46 Å Release Date: 2022-12-07 Classification: OXIDOREDUCTASE Ligands: FAD |
 |
Organism: Schizosaccharomyces pombe, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.81 Å Release Date: 2022-11-02 Classification: PROTEIN TRANSPORT |
 |
Crystal Structure Of Fusion Protein Of Human Tp53Inp2 Lir And Human Gabarap
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2022-11-02 Classification: PROTEIN TRANSPORT Ligands: IPA, PO4 |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2022-09-21 Classification: LIGASE |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.01 Å Release Date: 2022-06-22 Classification: SIGNALING PROTEIN Ligands: ANP |
 |
Organism: Homo sapiens
Method: SOLUTION NMR Release Date: 2022-06-08 Classification: STRUCTURAL PROTEIN |
 |
Crystal Structure Of Gabarap Complexed With The Tex264 Lir Phosphorylated At Ser271 And Ser272
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2022-03-30 Classification: SIGNALING PROTEIN |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.02 Å Release Date: 2022-03-30 Classification: SIGNALING PROTEIN Ligands: SO4 |

