Search Count: 390
All
Selected
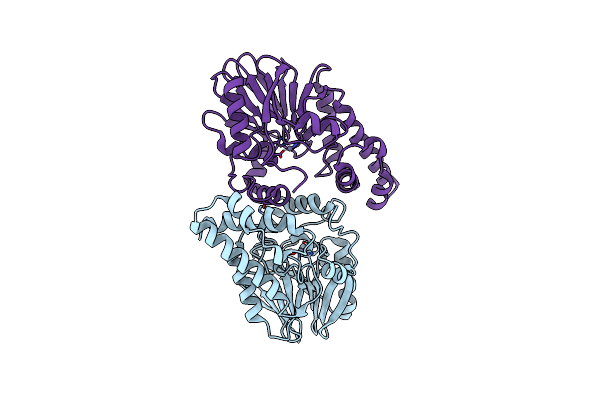 |
X-Ray Crystal Structure Of Chlorothalonil Dehalogenase: Analyzing The Catalytic Mechanism Of Hydrolytic Dehalogenation
Organism: Ochrobactrum sp. ctn-11
Method: X-RAY DIFFRACTION Resolution:1.96 Å Release Date: 2020-05-13 Classification: METAL BINDING PROTEIN Ligands: ZN |
 |
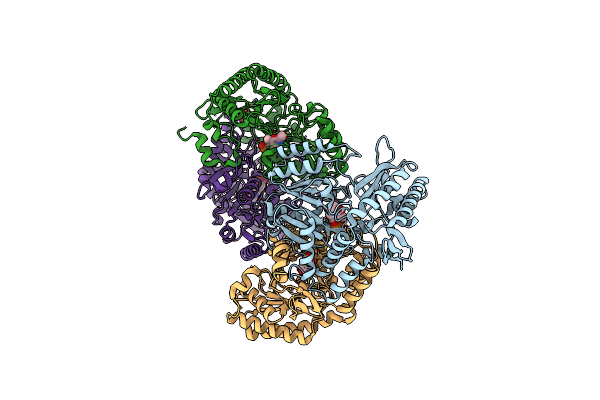
Crystal Structure Of Ribose-5-Phosphate Isomerase A From Ochrobactrum Sp. Csl1
Organism: Ochrobactrum sp.
Method: X-RAY DIFFRACTION Resolution:2.82 Å Release Date: 2019-04-17 Classification: ISOMERASE Ligands: PEG |
 |
Organism: Ochrobactrum anthropi
Method: X-RAY DIFFRACTION Resolution:1.87 Å Release Date: 2019-03-27 Classification: ISOMERASE Ligands: PLP, EDO |
 |
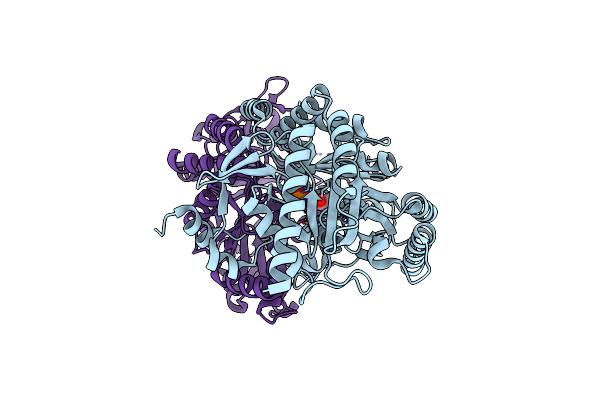
Structural And Functional Characterisation Of A Bacterial Laccase-Like Multi-Copper Oxidase Cueo From Lignin-Degrading Bacterium Ochrobactrum Sp. With Oxidase Activity Towards Lignin Model Compounds And Lignosulfonate
Organism: Ochrobactrum
Method: X-RAY DIFFRACTION Resolution:1.10 Å Release Date: 2018-04-11 Classification: OXIDOREDUCTASE Ligands: CU, EDO |
 |
Organism: Ochrobactrum anthropi (strain atcc 49188 / dsm 6882 / nctc 12168)
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2017-05-24 Classification: TRANSFERASE Ligands: PMP |
 |
Organism: Ochrobactrum anthropi (strain atcc 49188 / dsm 6882 / nctc 12168)
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2017-05-24 Classification: TRANSFERASE Ligands: PMP |
 |
Crystal Structure Of A Putative Periplasmic Solute Binding Protein (Ipr025997) From Ochrobactrum Anthropi Atcc49188 (Oant_2843, Target Efi-511085)
Organism: Ochrobactrum anthropi (strain atcc 49188 / dsm 6882 / nctc 12168)
Method: X-RAY DIFFRACTION Resolution:2.97 Å Release Date: 2015-06-10 Classification: SOLUTE-BINDING PROTEIN Ligands: HOH |
 |
Crystal Structure Of Sugar Transporter Oant_3817 From Ochrobactrum Anthropi, Target Efi-510528, With Bound Glucose
Organism: Ochrobactrum anthropi
Method: X-RAY DIFFRACTION Resolution:1.87 Å Release Date: 2014-08-27 Classification: TRANSPORT PROTEIN Ligands: GLC |
 |
Crystal Structure Of A Trap Periplasmic Solute Binding Protein From Ochrobactrum Anthropi (Oant_4429), Target Efi-510151, C-Termius Bound In Ligand Binding Pocket
Organism: Ochrobactrum anthropi
Method: X-RAY DIFFRACTION Resolution:1.30 Å Release Date: 2014-04-23 Classification: SOLUTE-BINDING PROTEIN |
 |
Crystal Structure Of A Trap Periplasmic Solute Binding Protein From Ochrobacterium Anthropi (Oant_3902), Target Efi-510153, With Bound L-Fuconate
Organism: Ochrobactrum anthropi
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2014-01-08 Classification: SOLUTE-BINDING PROTEIN Ligands: CL, LFC, PEG |
 |
Crystal Structure Of A Putative 4-Hydroxyproline Epimerase/3-Hydroxyproline Dehydratse From The Soil Bacterium Ochrobacterium Anthropi, Target Efi-506495, Disordered Loops
Organism: Ochrobactrum anthropi
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2013-05-01 Classification: ISOMERASE |
 |
Organism: Ochrobactrum
Method: X-RAY DIFFRACTION Resolution:1.29 Å Release Date: 2013-01-16 Classification: HYDROLASE |
 |
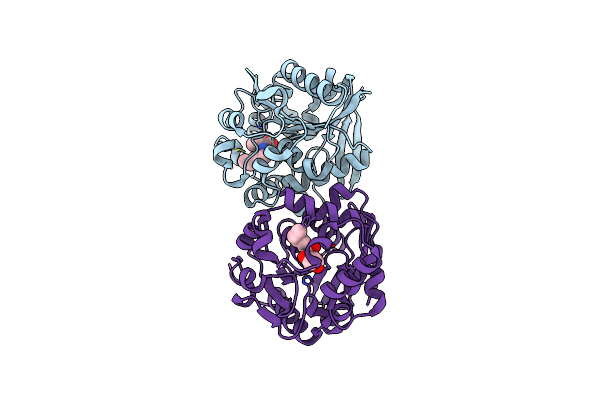
Crystal Structures Of N-Acyl Homoserine Lactonase Aidh S102G Mutant Complexed With N-Hexanoyl Homoserine Lactone
Organism: Ochrobactrum
Method: X-RAY DIFFRACTION Resolution:1.30 Å Release Date: 2013-01-16 Classification: HYDROLASE Ligands: HL6 |
 |
Crystal Structures Of N-Acyl Homoserine Lactonase Aidh E219G Mutant Complexed With N-Hexanoyl Homoserine
Organism: Ochrobactrum
Method: X-RAY DIFFRACTION Resolution:1.11 Å Release Date: 2013-01-16 Classification: HYDROLASE Ligands: C6L |
 |
Organism: Ochrobactrum
Method: X-RAY DIFFRACTION Resolution:1.35 Å Release Date: 2013-01-16 Classification: HYDROLASE |
 |
Crystal Structures Of N-Acyl Homoserine Lactonase Aidh Complexed With N-Butanoyl Homoserine
Organism: Ochrobactrum
Method: X-RAY DIFFRACTION Resolution:1.09 Å Release Date: 2013-01-16 Classification: HYDROLASE Ligands: C4L |
 |
Organism: Ochrobactrum
Method: X-RAY DIFFRACTION Resolution:1.35 Å Release Date: 2013-01-16 Classification: HYDROLASE Ligands: NI |
 |
Crystal Structure Of Ochrobactrum Anthropi Glutathione Transferase Cys10Ala Mutant With Glutathione Bound At The H-Site
Organism: Ochrobactrum anthropi
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2008-01-15 Classification: TRANSFERASE Ligands: SO4, GSH |
 |
Structure Of The Glutathione Transferase From Ochrobactrum Anthropi In Complex With Glutathione
Organism: Ochrobactrum anthropi
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2007-09-25 Classification: TRANSFERASE Ligands: SO4, GSH |
 |
The Crystal Structure Of D-Amino Acid Amidase From Ochrobactrum Anthropi Sv3 Complexed With L-Phenylalanine
Organism: Ochrobactrum anthropi
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2007-03-06 Classification: HYDROLASE Ligands: BA, PHE |

