Search Count: 16
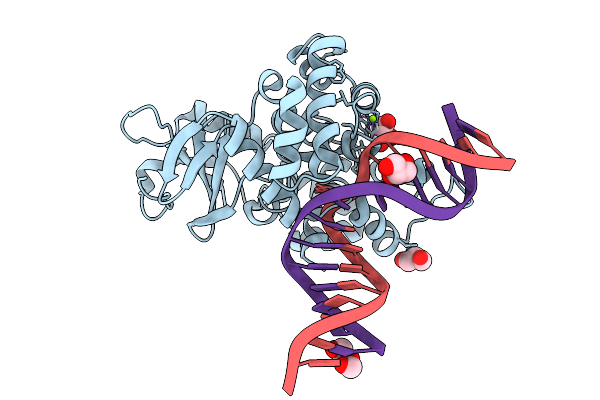 |
Crystal Structure Of Human 8-Oxoguanine Glycosylase K249H Mutant Bound To The Substrate 8-Oxoguanine Dna At Ph 8.0 Under 277 K
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-07-23 Classification: DNA/HYDROLASE Ligands: PEG, GOL, MG, NA |
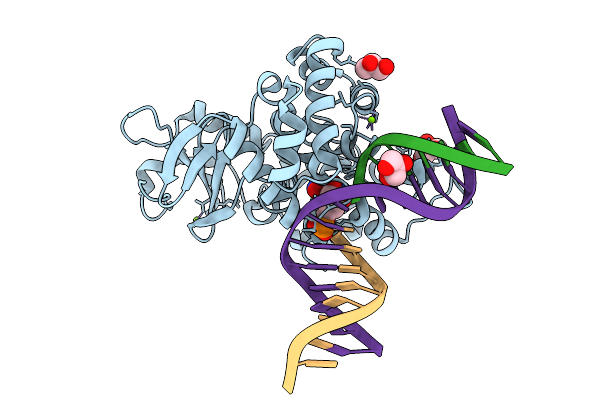 |
Crystal Structure Of Human 8-Oxoguanine Glycosylase K249H Mutant Bound To The Reaction Intermediate Derived From The Crystal Soaked Into The Solution At Ph 4.0 Under 277 K For 24 Hourss
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-07-23 Classification: DNA/HYDROLASE Ligands: A1LXK, MG, GOL |
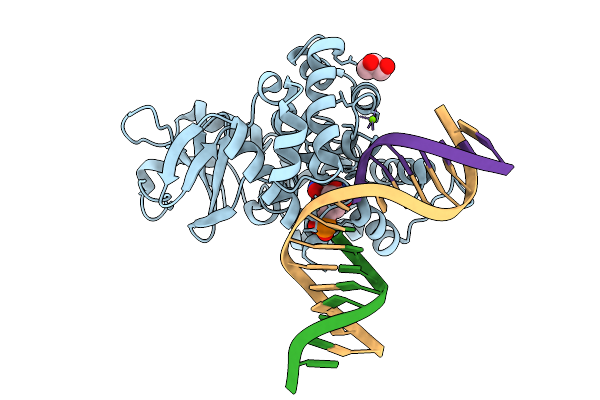 |
Crystal Structure Of Human 8-Oxoguanine Glycosylase K249H Mutant Bound To The Reaction Intermediate Derived From The Crystal Soaked Into The Solution At Ph 4.0 Under 277 K For 2.5 Hours
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-07-23 Classification: DNA/HYDROLASE Ligands: A1LXK, MG, GOL |
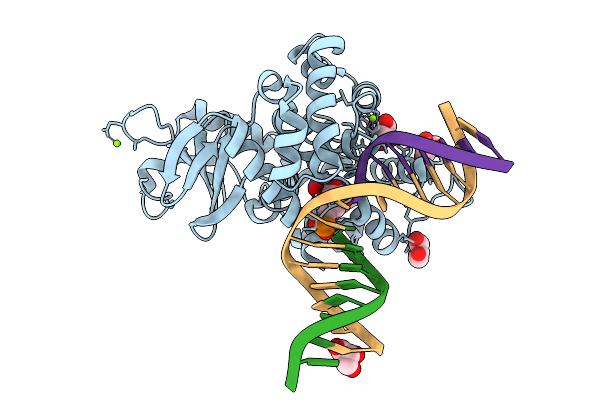 |
Crystal Structure Of Human 8-Oxoguanine Glycosylase K249H Mutant Bound To The Reaction Intermediate Derived From The Crystal Soaked Into The Solution At Ph 4.0 Under 298 K For 3 Weeks
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-07-23 Classification: DNA/HYDROLASE Ligands: A1LXK, GOL, MG, NA |
 |
Organism: Aspergillus oryzae
Method: X-RAY DIFFRACTION Release Date: 2020-05-13 Classification: OXIDOREDUCTASE Ligands: CU |
 |
Organism: Aspergillus oryzae
Method: X-RAY DIFFRACTION Release Date: 2020-05-13 Classification: OXIDOREDUCTASE Ligands: CU |
 |
Organism: Aspergillus oryzae
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2020-05-13 Classification: OXIDOREDUCTASE Ligands: NO3 |
 |
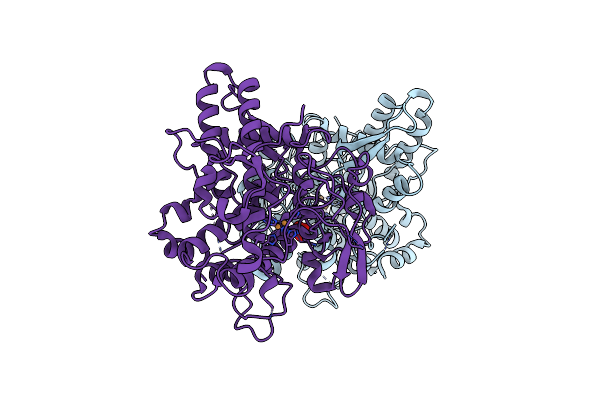
Aspergillus Oryzae Active-Tyrosinase Copper-Depleted C92A Mutant Complexed With L-Tyrosine
Organism: Aspergillus oryzae
Method: X-RAY DIFFRACTION Resolution:1.42 Å Release Date: 2020-05-13 Classification: OXIDOREDUCTASE Ligands: TYR, NO3 |
 |
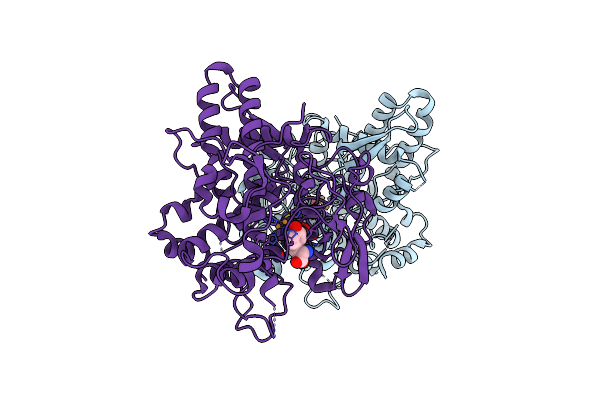
Organism: Aspergillus oryzae
Method: X-RAY DIFFRACTION Resolution:1.27 Å Release Date: 2020-05-13 Classification: OXIDOREDUCTASE Ligands: CU, NO3 |
 |
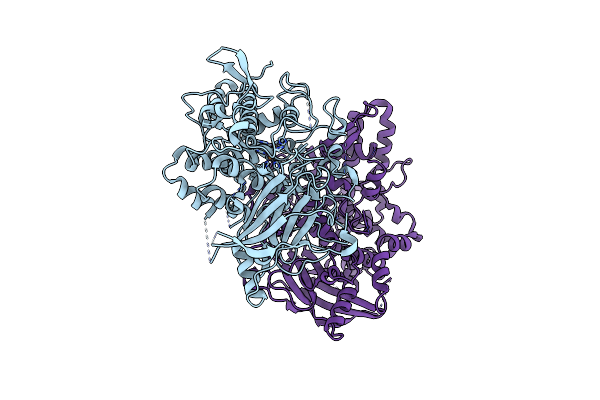
Aspergillus Oryzae Active-Tyrosinase Copper-Bound C92A Mutant Complexed With L-Tyrosine
Organism: Aspergillus oryzae
Method: X-RAY DIFFRACTION Release Date: 2020-05-13 Classification: OXIDOREDUCTASE Ligands: TYR, DAH, CU, NO3 |
 |
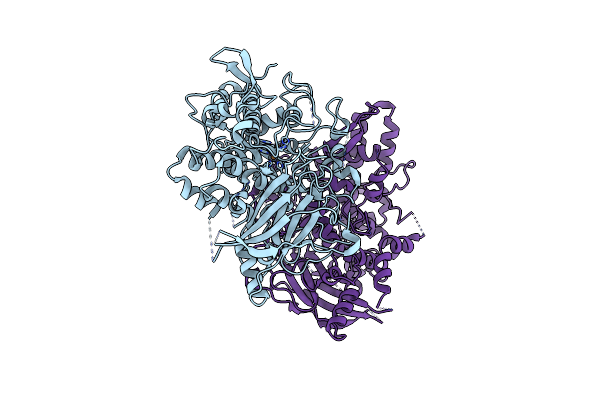
Organism: Aspergillus oryzae
Method: X-RAY DIFFRACTION Resolution:1.45 Å Release Date: 2020-05-13 Classification: OXIDOREDUCTASE Ligands: CU, PER |
 |
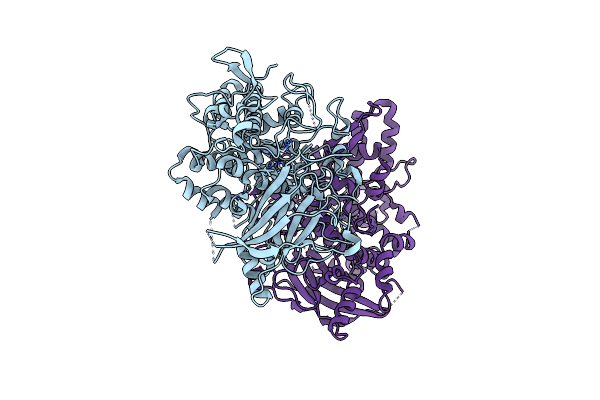
Radiation Damage In Aspergillus Oryzae Pro-Tyrosinase Oxygen-Bound C92A Mutant
Organism: Aspergillus oryzae
Method: X-RAY DIFFRACTION Resolution:1.54 Å Release Date: 2020-05-13 Classification: OXIDOREDUCTASE Ligands: CU, PER |
 |
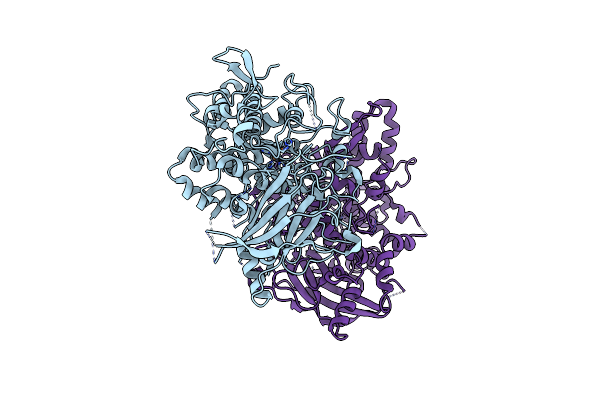
Organism: Aspergillus oryzae
Method: X-RAY DIFFRACTION Resolution:1.44 Å Release Date: 2020-05-13 Classification: OXIDOREDUCTASE Ligands: CU, PER |
 |
Radiation Damage In Aspergillus Oryzae Pro-Tyrosinase Oxygen-Bound C92A/H103F Mutant
Organism: Aspergillus oryzae
Method: X-RAY DIFFRACTION Resolution:1.56 Å Release Date: 2020-05-13 Classification: OXIDOREDUCTASE Ligands: CU, PER |
 |
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:3.45 Å Release Date: 2014-03-05 Classification: PROTON TRANSPORT |
 |
Crystal Structure Of A Parallel Coiled-Coil Dimerization Domain From The Voltage-Gated Proton Channel
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:1.45 Å Release Date: 2012-05-30 Classification: MEMBRANE PROTEIN |

