Search Count: 12
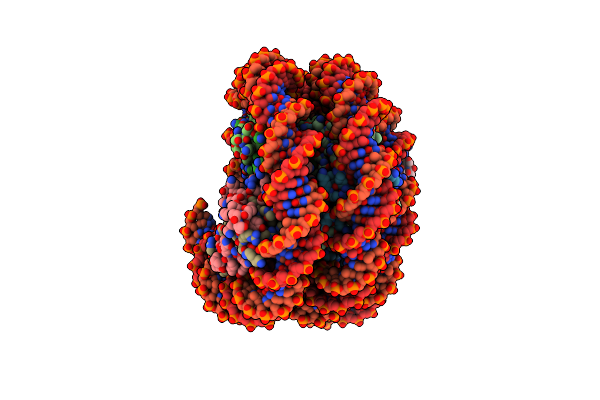 |
Structure Of A Hexasome-Nucleosome Complex With A Dyad-To-Dyad Distance Of 103 Bp.
Organism: Xenopus laevis, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2024-09-18 Classification: DNA BINDING PROTEIN |
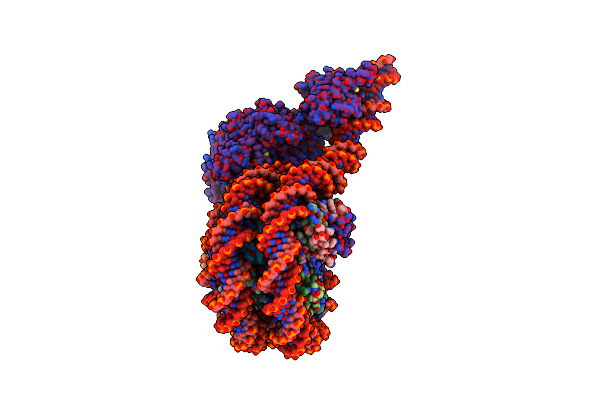 |
Structure Of Chd1 Bound To A Hexasome-Nucleosome Complex With A Dyad-To-Dyad Distance Of 103 Bp.
Organism: Xenopus laevis, Synthetic construct, Saccharomyces cerevisiae s288c
Method: ELECTRON MICROSCOPY Release Date: 2024-09-18 Classification: DNA BINDING PROTEIN Ligands: ADP, BEF, MG |
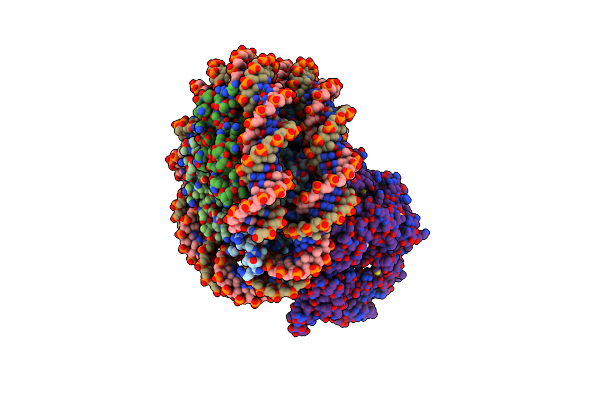 |
Structure Of Chd1 Bound To A Dinucleosome With A Dyad-To-Dyad Distance Of 103 Bp.
Organism: Xenopus laevis, Synthetic construct, Saccharomyces cerevisiae s288c
Method: ELECTRON MICROSCOPY Release Date: 2024-09-18 Classification: DNA BINDING PROTEIN Ligands: ADP, BEF, MG |
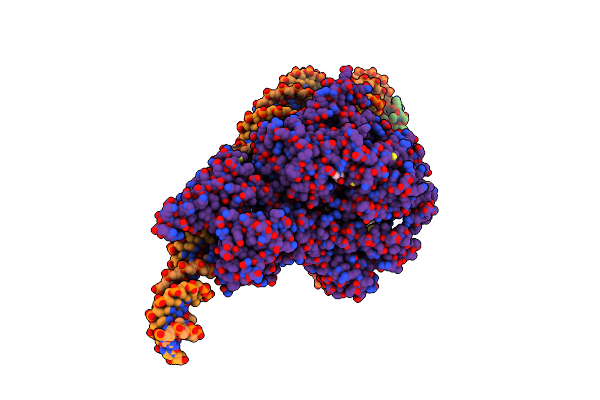 |
Structure Of A Mononucleosome Bound By One Copy Of Chd1 With The Dbd On The Exit-Side Dna.
Organism: Xenopus laevis, Synthetic construct, Saccharomyces cerevisiae s288c
Method: ELECTRON MICROSCOPY Release Date: 2024-09-18 Classification: DNA BINDING PROTEIN Ligands: ADP, BEF, MG |
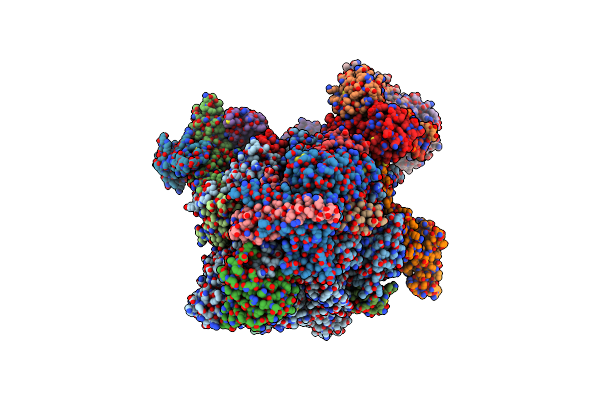 |
Rna Polymerase Ii Core Initially Transcribing Complex With An Ordered Rna Of 8 Nt
Organism: Homo sapiens, Unidentified adenovirus, Sus scrofa
Method: ELECTRON MICROSCOPY Release Date: 2024-04-17 Classification: TRANSCRIPTION Ligands: ZN, MG |
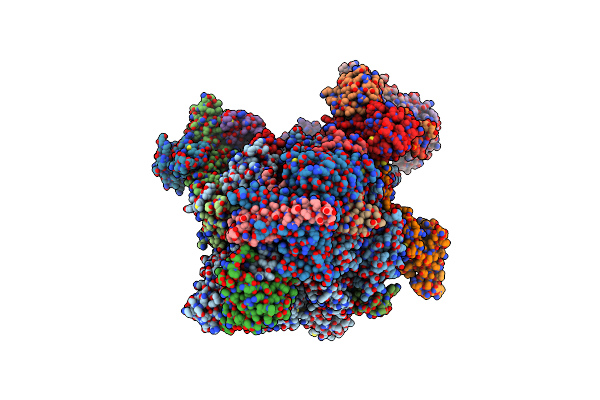 |
Rna Polymerase Ii Core Initially Transcribing Complex With An Ordered Rna Of 10 Nt
Organism: Homo sapiens, Unidentified adenovirus, Sus scrofa
Method: ELECTRON MICROSCOPY Release Date: 2024-04-17 Classification: TRANSCRIPTION Ligands: ZN, MG |
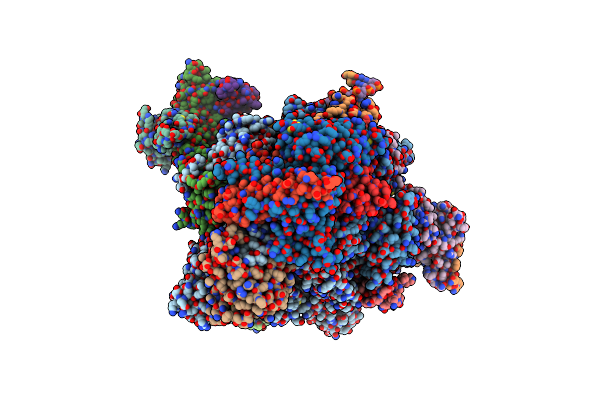 |
Rna Polymerase Ii Early Elongation Complex Bound To Tfiie And Tfiif - State B (Composite Structure)
Organism: Homo sapiens, Unidentified adenovirus, Sus scrofa
Method: ELECTRON MICROSCOPY Release Date: 2024-04-17 Classification: TRANSCRIPTION Ligands: ZN, MG |
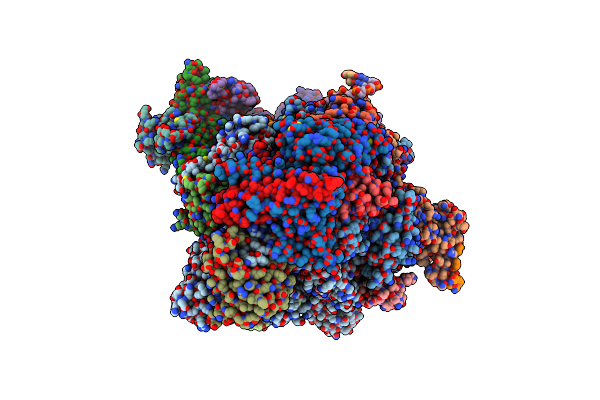 |
Rna Polymerase Ii Early Elongation Complex Bound To Tfiie And Tfiif - State A (Composite Structure)
Organism: Homo sapiens, Unidentified adenovirus, Sus scrofa
Method: ELECTRON MICROSCOPY Release Date: 2024-04-17 Classification: TRANSCRIPTION Ligands: ZN, MG |
 |
Rna Polymerase Ii Core Initially Transcribing Complex With An Ordered Rna Of 12 Nt
Organism: Homo sapiens, Unidentified adenovirus, Sus scrofa
Method: ELECTRON MICROSCOPY Release Date: 2024-04-10 Classification: TRANSCRIPTION Ligands: ZN, MG |
 |
Crystal Structure Of The Arp4-N-Actin(Apo-State) Heterodimer Bound By A Nanobody
Organism: Saccharomyces cerevisiae (strain atcc 204508 / s288c), Vicugna pacos, Trichoplusia ni
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2018-08-22 Classification: HYDROLASE Ligands: CA, ATP |
 |
Crystal Structure Of The Arp4-N-Actin(Atp-State) Heterodimer Bound By A Nanobody
Organism: Saccharomyces cerevisiae (strain atcc 204508 / s288c), Vicugna pacos, Trichoplusia ni
Method: X-RAY DIFFRACTION Resolution:3.40 Å Release Date: 2018-08-22 Classification: HYDROLASE Ligands: CA, ATP |
 |
Organism: Saccharomyces cerevisiae (strain atcc 204508 / s288c)
Method: X-RAY DIFFRACTION Resolution:4.00 Å Release Date: 2018-08-22 Classification: HYDROLASE Ligands: CA, ATP, LAR |

