Search Count: 542
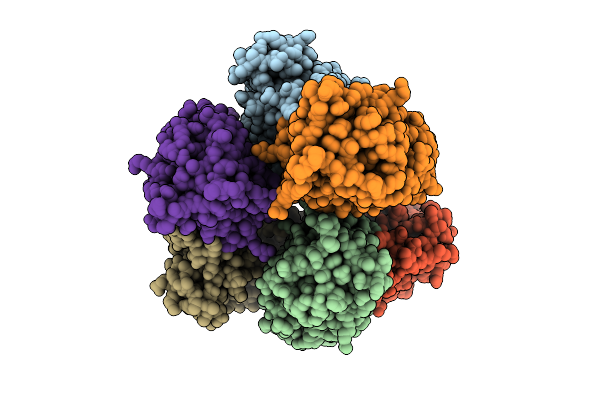 |
Crystal Structure Of Non-Haem Diiron Azetidine Synthase From Streptomyces Cacaoi Var. Asoensis Complexed With Iron, L-Isoleucine And Molecular Oxygen
Organism: Streptomyces cacaoi
Method: X-RAY DIFFRACTION Release Date: 2025-12-24 Classification: METAL BINDING PROTEIN Ligands: ILE, FE2, OXY |
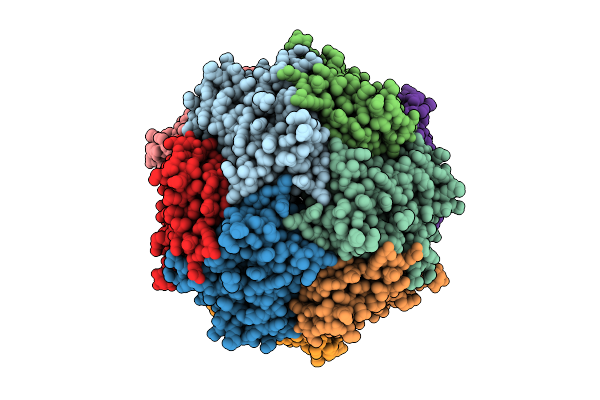 |
Structure Of Thioferritin (Pfdpsl) With Ferrihydrite Growth At A Single Three-Fold Pore.
Organism: Pyrococcus furiosus
Method: ELECTRON MICROSCOPY Release Date: 2025-10-29 Classification: METAL BINDING PROTEIN Ligands: FE, OXY, O |
 |
Crystal Structure Of Dehaloperoxidase B In Complex With Substrate 1,4-Cyclohexadiene
Organism: Amphitrite ornata
Method: X-RAY DIFFRACTION Release Date: 2025-08-13 Classification: OXIDOREDUCTASE Ligands: HEM, A1ADJ, GOL, SO4, OXY |
 |
Organism: Albifimbria verrucaria
Method: X-RAY DIFFRACTION Release Date: 2025-08-06 Classification: OXIDOREDUCTASE Ligands: CU, ATP, OXY, CL |
 |
Organism: Albifimbria verrucaria
Method: X-RAY DIFFRACTION Release Date: 2025-08-06 Classification: OXIDOREDUCTASE Ligands: CU, SIN, OXY, PG4, PGR, NAD, GOL |
 |
Organism: Albifimbria verrucaria
Method: X-RAY DIFFRACTION Release Date: 2025-08-06 Classification: OXIDOREDUCTASE Ligands: CU, OXY, GOL, SIN, CL |
 |
Organism: Albifimbria verrucaria
Method: X-RAY DIFFRACTION Release Date: 2025-08-06 Classification: OXIDOREDUCTASE Ligands: CU, OXY, GOL, PGE, MLA, CL |
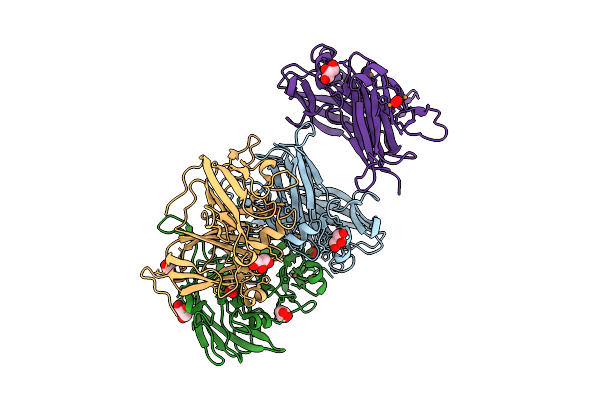 |
Mutant H286T Crystal Structure Of Two-Domain Bacterial Laccase From The Actinobacterium Streptomyces Carpinensis Vkm Ac-1300
Organism: Streptomyces carpinensis
Method: X-RAY DIFFRACTION Release Date: 2025-08-06 Classification: OXIDOREDUCTASE Ligands: CU, OXY, GOL |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-07-02 Classification: ENDOCYTOSIS Ligands: NAG, CA, HEM, OXY |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-07-02 Classification: ENDOCYTOSIS Ligands: NAG, CA, HEM, OXY |
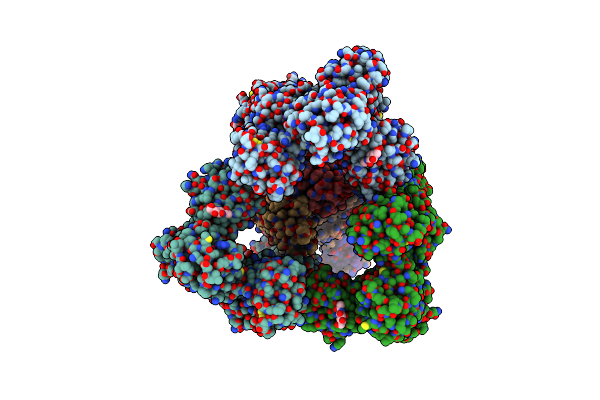 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-06-18 Classification: ENDOCYTOSIS Ligands: NAG, CA, HEM, OXY |
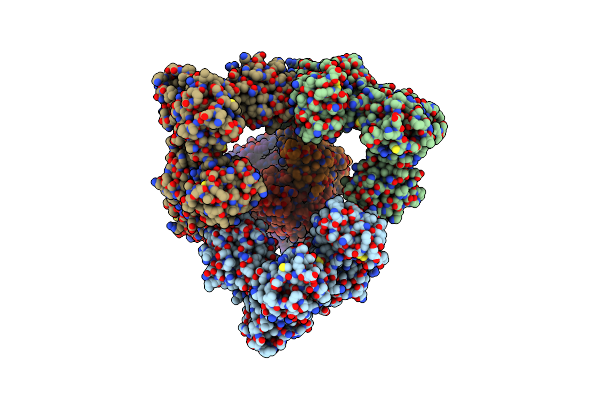 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-06-18 Classification: ENDOCYTOSIS Ligands: CA, HEM, OXY |
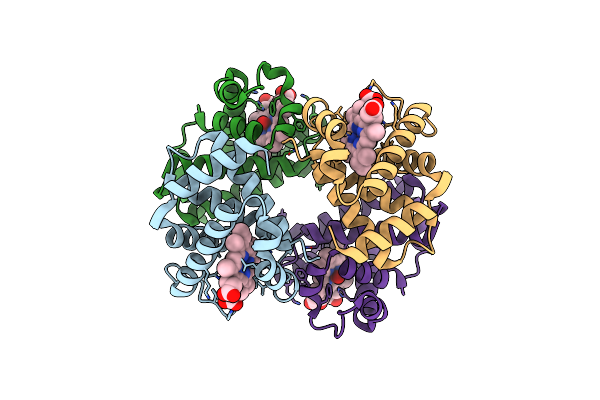 |
Human Oxyhb (C1 Symmetry) Obtained Using The Spt Labtech Chameleon In The Presence Of 25 Um Sodium Dithionite Under Al'S Oil
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-04-30 Classification: OXYGEN BINDING Ligands: HEM, OXY |
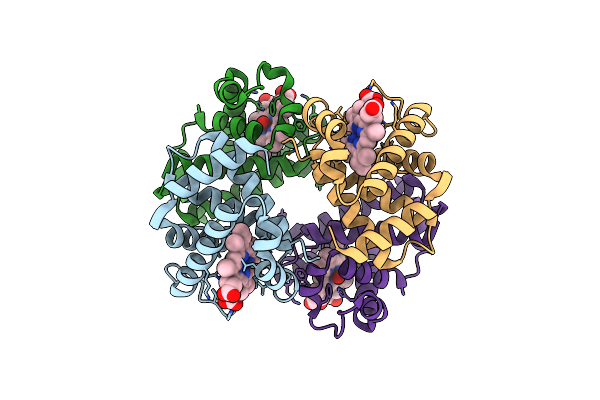 |
Human Oxyhb (C2 Symmetry) Obtained Using The Spt Labtech Chameleon In The Presence Of 25 Um Sodium Dithionite Under Al'S Oil
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-04-30 Classification: OXYGEN BINDING Ligands: HEM, OXY |
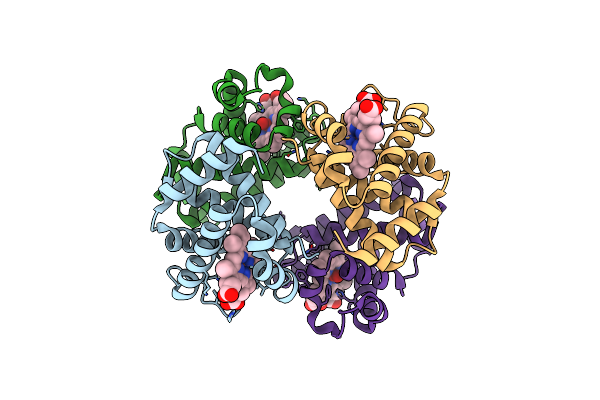 |
Human Oxyhb (C1 Symmetry) Obtained Using The Spt Labtech Chameleon In The Presence Of 5 Mm Sodium Dithionite Under Al'S Oil
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-04-30 Classification: OXYGEN BINDING Ligands: HEM, OXY |
 |
Human Oxyhb (C2 Symmetry) Obtained Using The Spt Labtech Chameleon In The Presence Of 5 Mm Sodium Dithionite Under Al'S Oil
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-04-30 Classification: OXYGEN BINDING Ligands: HEM, OXY |
 |
Human Oxyhb (C1 Symmetry) Obtained Using The Spt Labtech Chameleon In The Presence Of 20 Mm Sodium Dithionite Under Al'S Oil
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-04-30 Classification: OXYGEN BINDING Ligands: HEM, OXY |
 |
Crystal Structure Of The Dyp-Type Peroxidase Pross Variant From Pseudomonas Putida
Organism: Pseudomonas putida
Method: X-RAY DIFFRACTION Release Date: 2025-04-30 Classification: OXIDOREDUCTASE Ligands: OXY, HEM, CL |
 |
Synchrotron X-Ray Crystal Structure Of Oxygen-Bound F87A/F393H P450Bm3 With Decoy C7Prophe (N-Enanthyl-L-Prolyl-L-Phenylalanine) And Substrate Styrene At 2 Mgy X-Ray Dose
Organism: Priestia megaterium nbrc 15308 = atcc 14581
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2025-03-26 Classification: OXIDOREDUCTASE Ligands: HEM, SYN, OXY, D0L, GOL |
 |
Organism: Phytophthora sojae
Method: X-RAY DIFFRACTION Resolution:2.12 Å Release Date: 2025-03-12 Classification: OXIDOREDUCTASE Ligands: NAG, FAD, OXY, MAN |

