Search Count: 98,001
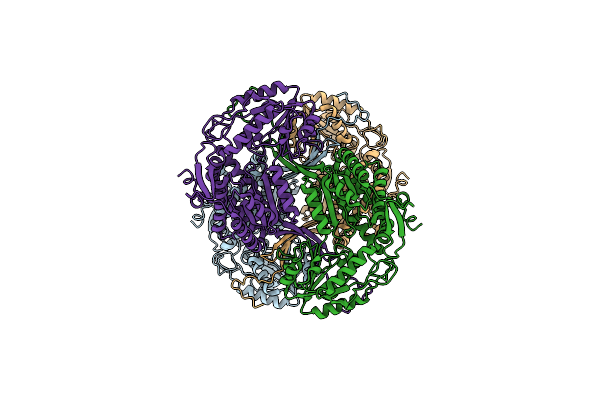 |
Cryoem Structure Of Aldehyde Dehydrogenase From Francisella Tularensis Subsp. Tularensis At 3.03A Resolution
Organism: Francisella tularensis subsp. tularensis
Method: ELECTRON MICROSCOPY Resolution:3.03 Å Release Date: 2026-01-28 Classification: OXIDOREDUCTASE |
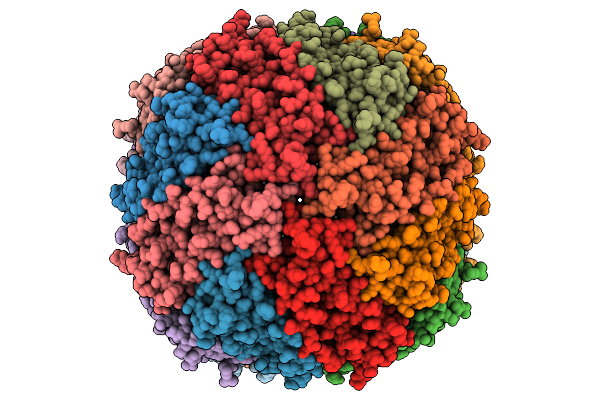 |
Cryo-Et Subtomogram-Averaged Structure Of Mouse Heavy-Chain Apoferritin Resolved At 2.71 Angstroms
Organism: Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2026-01-28 Classification: OXIDOREDUCTASE Ligands: MG |
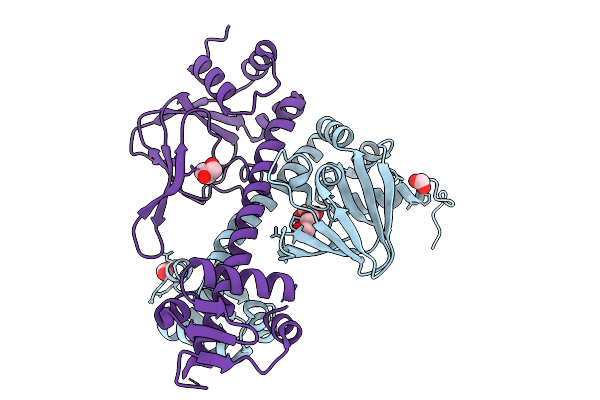 |
Crystal Structure Of The Transcriptional Regulator Hcpr From Porphyromonas Gingivalis
Organism: Porphyromonas gingivalis
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2026-01-28 Classification: TRANSCRIPTION Ligands: GOL, FMT |
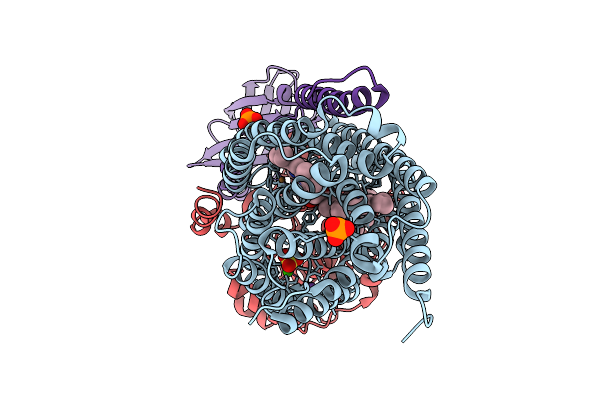 |
Organism: Stutzerimonas stutzeri atcc 14405 = ccug 16156
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2026-01-28 Classification: OXIDOREDUCTASE Ligands: HEM, CU, CA, PO4, NA, SO4, HEC |
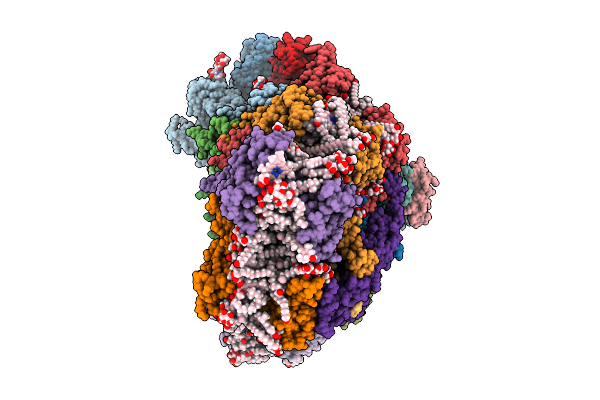 |
Cryo-Em Structure Of The Chromera Velia Psi Supercomplex At 1.84 Angstrom Resolution
Organism: Chromera velia
Method: ELECTRON MICROSCOPY Resolution:1.84 Å Release Date: 2026-01-28 Classification: PHOTOSYNTHESIS Ligands: FE, CLA, CL0, SF4, PQN, XAT, LMG, DGD, LMT, BCR, AV0, A1L6D, SQD, A1I05 |
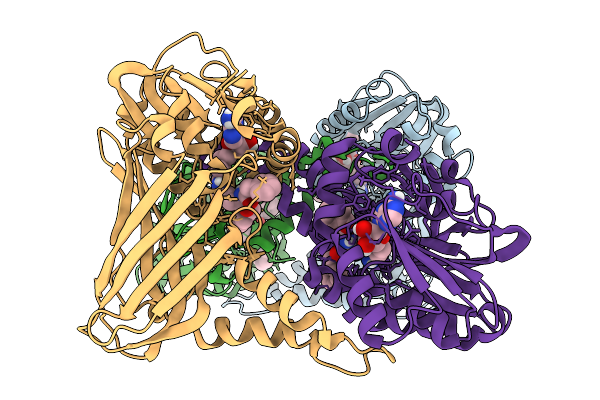 |
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:2.22 Å Release Date: 2026-01-28 Classification: TRANSFERASE Ligands: A1IZI |
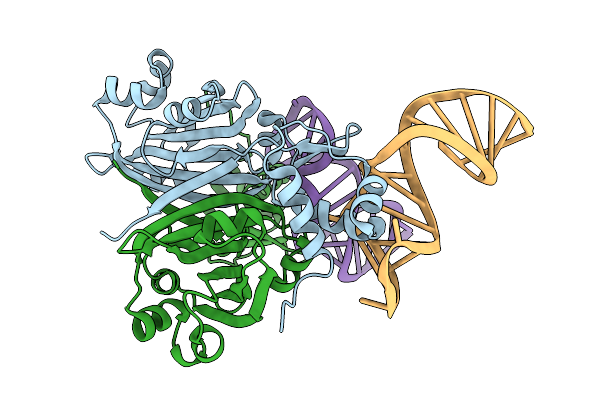 |
Crystal Structure Of Dasr In Complex With A Synthetic Dasr-Binding Rna Aptamer
Organism: Streptomyces coelicolor, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.02 Å Release Date: 2026-01-28 Classification: RNA BINDING PROTEIN Ligands: BR |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.63 Å Release Date: 2026-01-28 Classification: METAL BINDING PROTEIN Ligands: FE, MG, CL |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.73 Å Release Date: 2026-01-28 Classification: METAL BINDING PROTEIN Ligands: FE, CL, MG |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.63 Å Release Date: 2026-01-28 Classification: METAL BINDING PROTEIN Ligands: FE, CL, MG |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.58 Å Release Date: 2026-01-28 Classification: METAL BINDING PROTEIN Ligands: FE, MG, CL |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2026-01-28 Classification: METAL BINDING PROTEIN Ligands: FE, MG, CL |
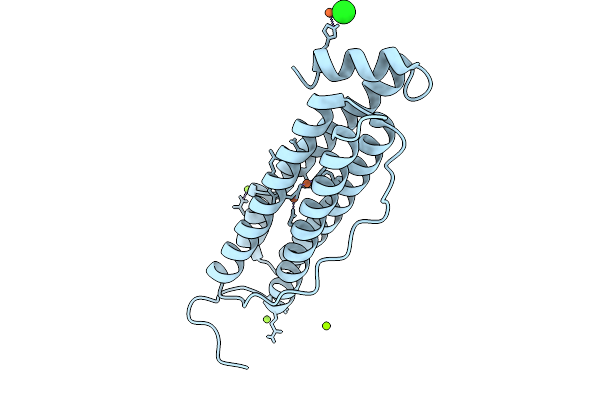 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.76 Å Release Date: 2026-01-28 Classification: METAL BINDING PROTEIN Ligands: FE, CL, MG |
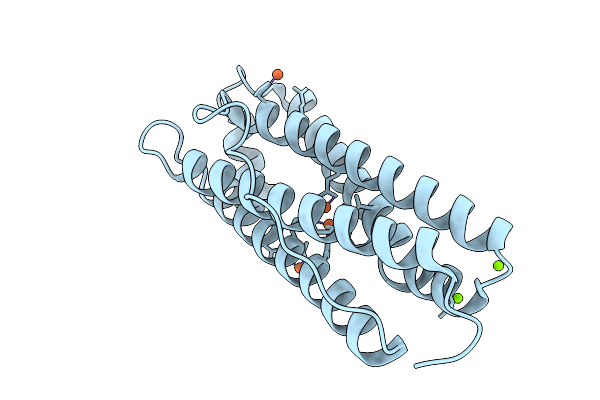 |
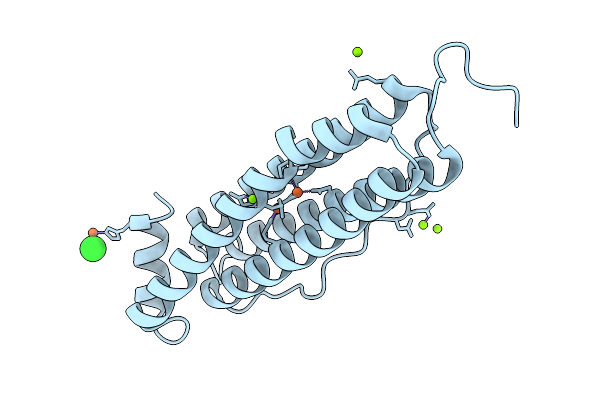
Iron Loaded Human Mitochondrial Ferritin D131N Mutant 20 Minute Oxygen Soak
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.72 Å Release Date: 2026-01-28 Classification: METAL BINDING PROTEIN Ligands: FE, MG, CL |
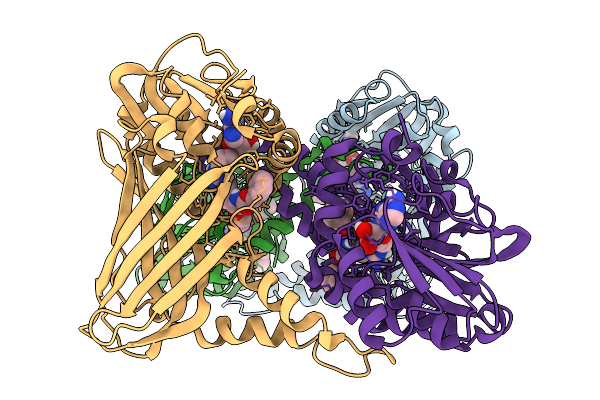 |
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:2.39 Å Release Date: 2026-01-28 Classification: TRANSCRIPTION Ligands: A1IZ2 |
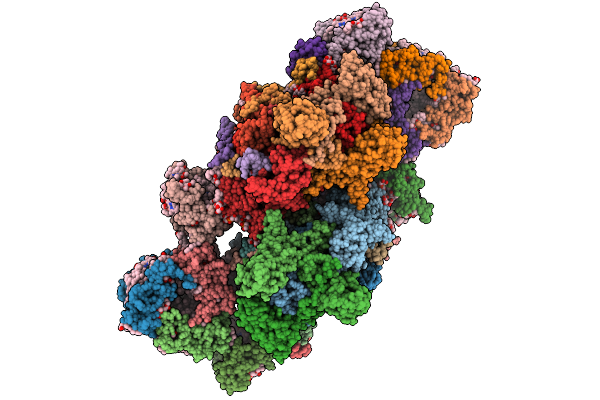 |
Structure Of Unstacked C2S2-Type Psii-Lhcii Supercomplex From Pisum Sativum
|
 |
The Apo-Mtrex1 Crystal Structure For Soaking Experiments (Soaking Condition 1)
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2026-01-28 Classification: HYDROLASE Ligands: ACT |
 |
The Apo-Mtrex1 Crystal Structure For Soaking Experiments (Soaking Condition 2)
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2026-01-28 Classification: HYDROLASE Ligands: ACT |
 |
The Apo-Mtrex1 Crystal Structure For Soaking Experiments (Soaking Condition 3)
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2026-01-28 Classification: HYDROLASE Ligands: GOL |
 |
The Mtrex1-Nsc 37203 Complex Structure By Co-Crystallization (Nsc 37203 Complex 1)
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:1.91 Å Release Date: 2026-01-28 Classification: HYDROLASE Ligands: DMS, LI, EPE, SO4, A1L5X |

