Search Count: 40,254
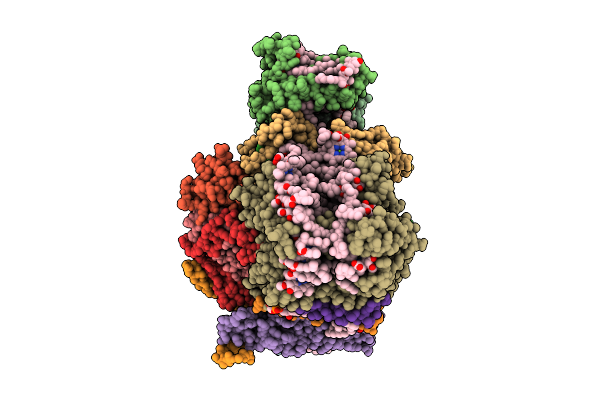 |
Structure Of The Wild-Type Psi-5Vcpi Supercomplex In Nannochloropsis Oceanica
Organism: Nannochloropsis oceanica strain imet1
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: PHOTOSYNTHESIS Ligands: XAT, A1L1G, CLA, SQD, DGD, LMG, A1L1F, PQN, LHG, BCR, SF4 |
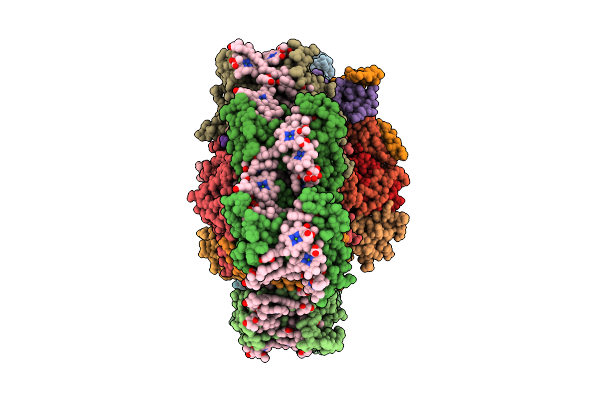 |
Structure Of The Canthaxanthin Mutant Psi-9Vcpi Supercomplex In Nannochloropsis Oceanica
Organism: Nannochloropsis oceanica strain imet1
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: PHOTOSYNTHESIS Ligands: XAT, A1L1G, CLA, SQD, A1L1F, 45D, LHG, DGD, LMG, PQN, BCR, SF4 |
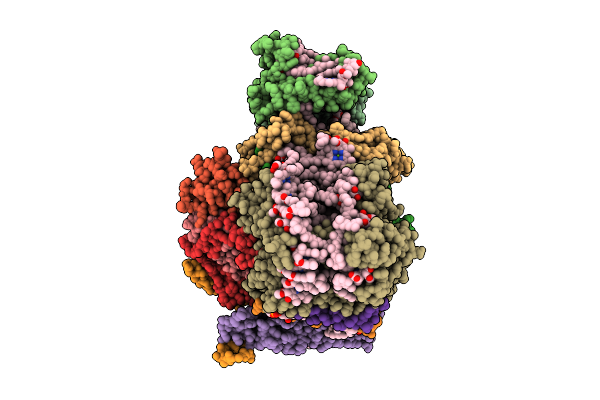 |
Structure Of The Canthaxanthin Mutant Psi-5Vcpi Supercomplex In Nannochloropsis Oceanica
Organism: Nannochloropsis oceanica strain imet1
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: PHOTOSYNTHESIS Ligands: XAT, A1L1G, CLA, SQD, DGD, LMG, A1L1F, PQN, LHG, BCR, SF4 |
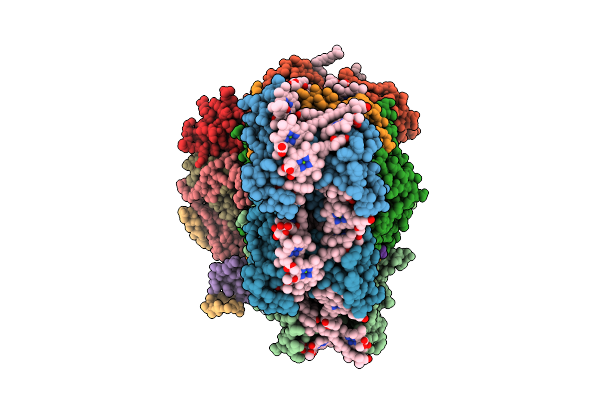 |
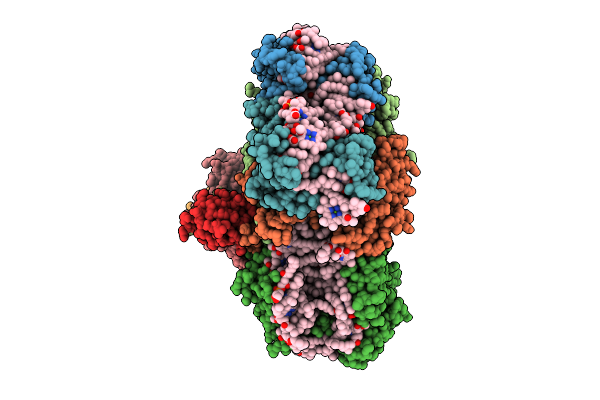
Structure Of The Canthaxanthin Mutant Psi-4Vcpi Supercomplex In Nannochloropsis Oceanica
Organism: Nannochloropsis oceanica strain imet1
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: PHOTOSYNTHESIS Ligands: A1L1G, A1L1F, XAT, 45D, LHG, CLA, DGD, SQD, PQN, BCR, SF4, LMG |
 |
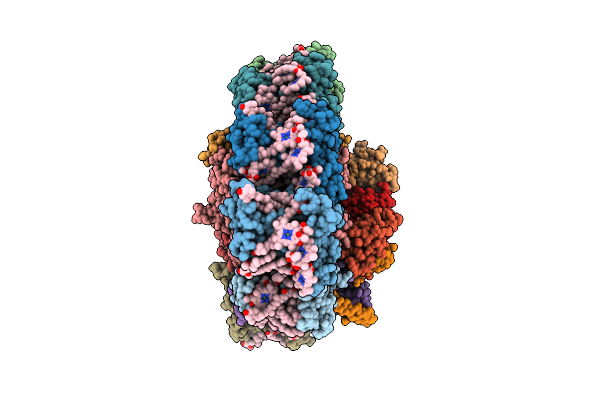
Structure Of The Canthaxanthin Mutant Psi-3Vcpi Supercomplex In Nannochloropsis Oceanica
Organism: Nannochloropsis oceanica strain imet1
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: PHOTOSYNTHESIS Ligands: XAT, A1L1G, CLA, SQD, DGD, A1L1F, PQN, LHG, BCR, SF4, LMG |
 |
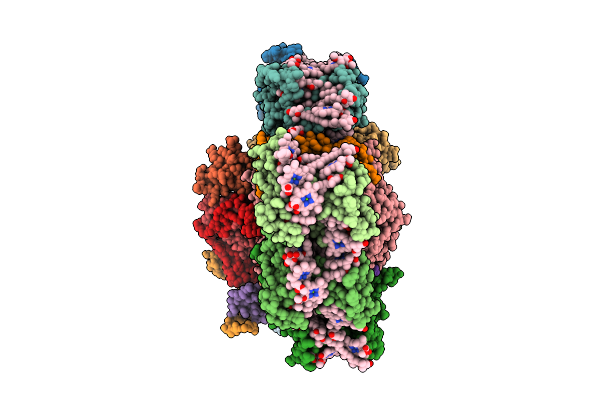
Structure Of The Astaxanthin Mutant Psi-9Vcpi Supercomplex In Nannochloropsis Oceanica
Organism: Nannochloropsis oceanica strain imet1
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: PHOTOSYNTHESIS Ligands: XAT, A1L1G, CLA, SQD, A1L1F, LHG, DGD, LMG, PQN, BCR, SF4 |
 |
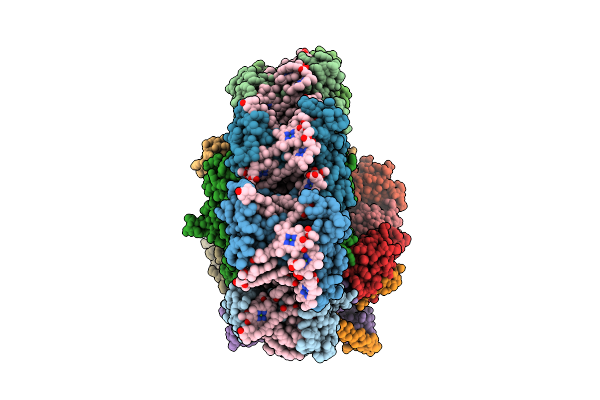
Structure Of The Astaxanthin Mutant Psi-7Vcpi Supercomplex In Nannochloropsis Oceanica
Organism: Nannochloropsis oceanica strain imet1
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: PHOTOSYNTHESIS Ligands: XAT, A1L1G, CLA, SQD, A1L1F, LHG, DGD, PQN, BCR, SF4, LMG |
 |
Structure Of The Astaxanthin Mutant Psi-5Vcpi Supercomplex In Nannochloropsis Oceanica
Organism: Nannochloropsis oceanica strain imet1
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: PHOTOSYNTHESIS Ligands: XAT, A1L1G, CLA, SQD, DGD, LMG, A1L1F, PQN, LHG, BCR, SF4 |
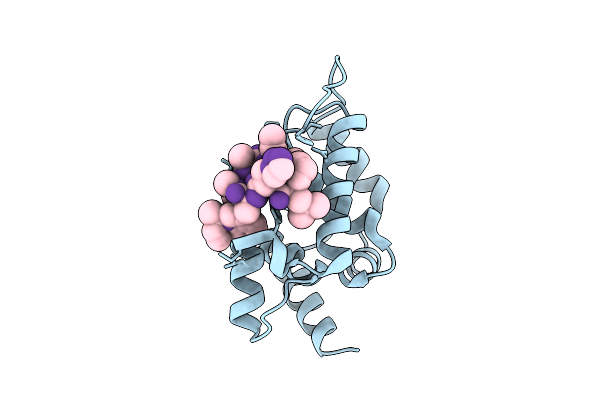 |
M. Tuberculosis Clpc1-Ntd Complexed With A Click Chemistry Analog Of Rufomycin
Organism: Mycobacterium tuberculosis, Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-12-10 Classification: ANTIBIOTIC |
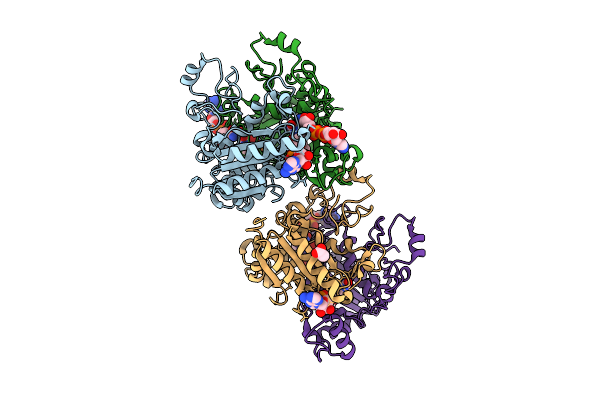 |
Xfel Structure Of Hnqo1 Mixed With Nadh In An Extended Orientation At 0.3 S
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-12-10 Classification: OXIDOREDUCTASE Ligands: NAD, FAD, ACT |
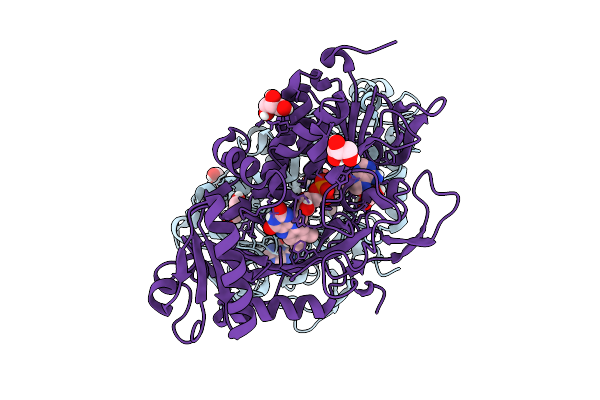 |
Organism: Parastagonospora nodorum sn15
Method: X-RAY DIFFRACTION Release Date: 2025-12-10 Classification: OXIDOREDUCTASE Ligands: FAD, GOL |
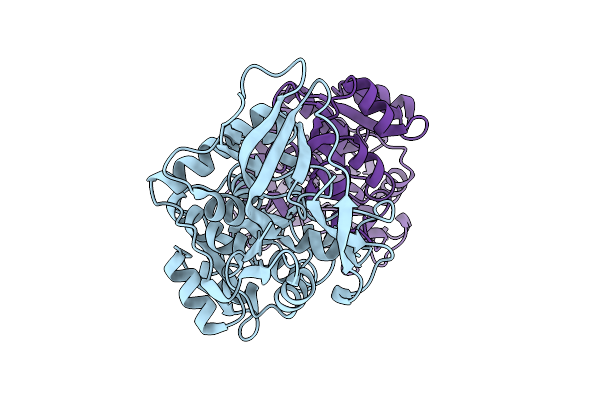 |
Organism: Rouxiella badensis
Method: X-RAY DIFFRACTION Release Date: 2025-12-10 Classification: OXIDOREDUCTASE |
 |
Organism: Mesorhizobium metallidurans
Method: X-RAY DIFFRACTION Release Date: 2025-12-10 Classification: OXIDOREDUCTASE Ligands: P6G, PEG, EPE, NA, TRS |
 |
Organism: Escherichia coli k-12
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: ELECTRON TRANSPORT Ligands: FE2, FES, HCI |
 |
Organism: Escherichia coli k-12
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: STRUCTURAL PROTEIN Ligands: NAP, ACO, MG |
 |
Organism: Acinetobacter baumannii
Method: X-RAY DIFFRACTION Release Date: 2025-12-10 Classification: OXIDOREDUCTASE Ligands: FAD |
 |
Organism: Escherichia coli k-12
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: STRUCTURAL PROTEIN Ligands: NAP, MG |
 |
Organism: Escherichia coli k-12
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: STRUCTURAL PROTEIN Ligands: NAP, MG |
 |
Crystal Structure Of Human Prostaglandin Reductase 1 (Ptgr1) In Complex With Nadp
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-12-10 Classification: OXIDOREDUCTASE Ligands: CL, NAP, PG6 |
 |
Crystal Structure Of Human Prostaglandin Reductase 1 (Ptgr1) In Complex With Nadp And Indomethacin
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-12-10 Classification: OXIDOREDUCTASE Ligands: ACT, IMN, NAP, GOL |

