Search Count: 129
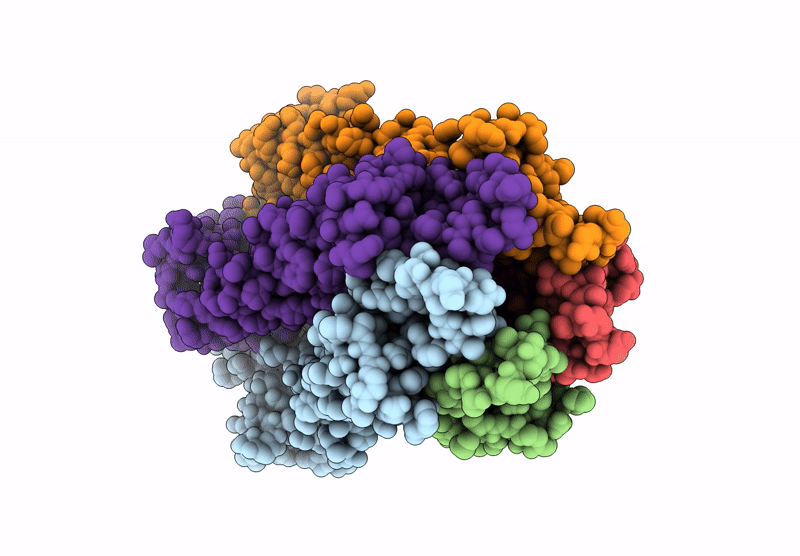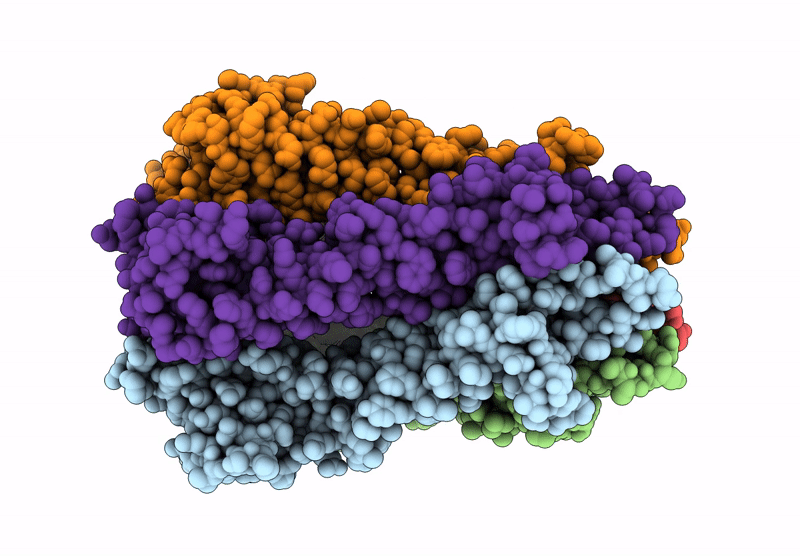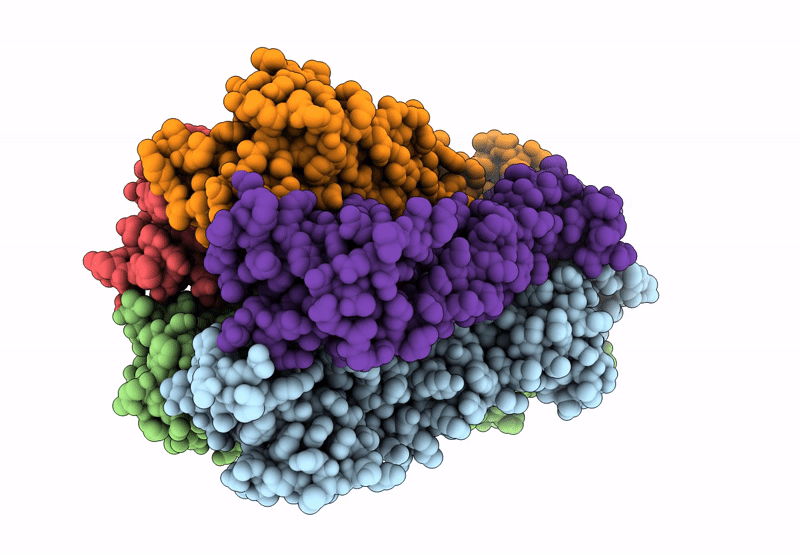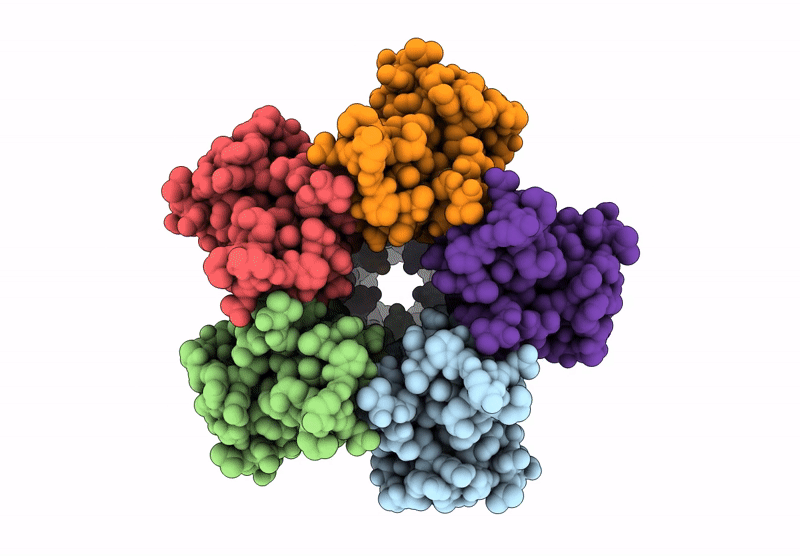 |
Organism: Acinetobacter baumannii
Method: ELECTRON MICROSCOPY Resolution:3.02 Å Release Date: 2025-01-22 Classification: MEMBRANE PROTEIN |
 |
Organism: Acinetobacter baumannii
Method: ELECTRON MICROSCOPY Resolution:3.34 Å Release Date: 2025-01-15 Classification: MEMBRANE PROTEIN |
 |
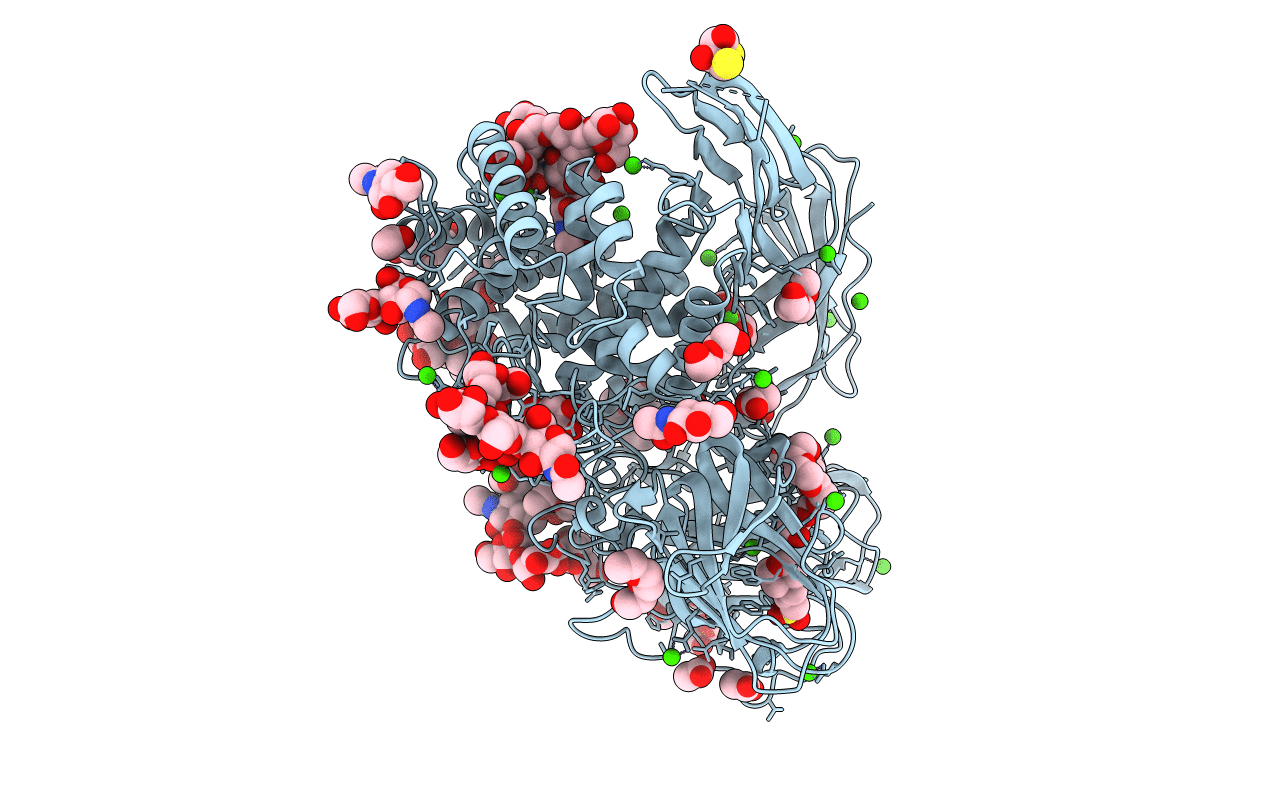
Solvent Organization In Ultrahigh-Resolution Protein Crystal Structure At Room Temperature
Organism: Crambe hispanica subsp. abyssinica
Method: X-RAY DIFFRACTION Resolution:0.70 Å Release Date: 2024-09-04 Classification: PLANT PROTEIN Ligands: EOH |
 |
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2022-11-30 Classification: PROTON TRANSPORT Ligands: HEM, HEO, CU, 3PE |
 |
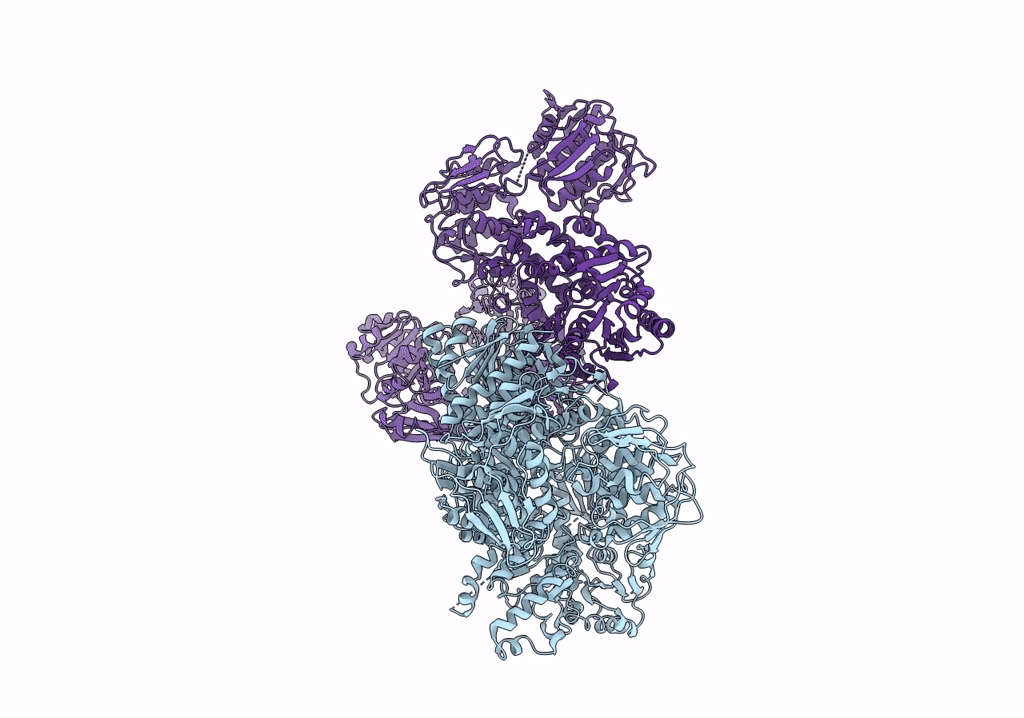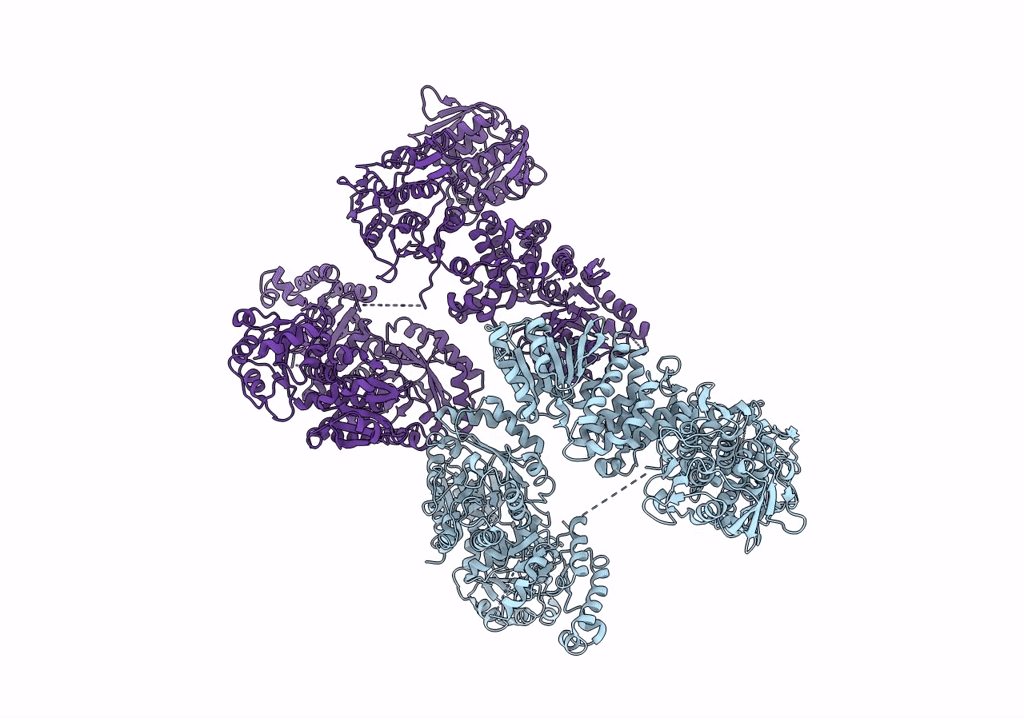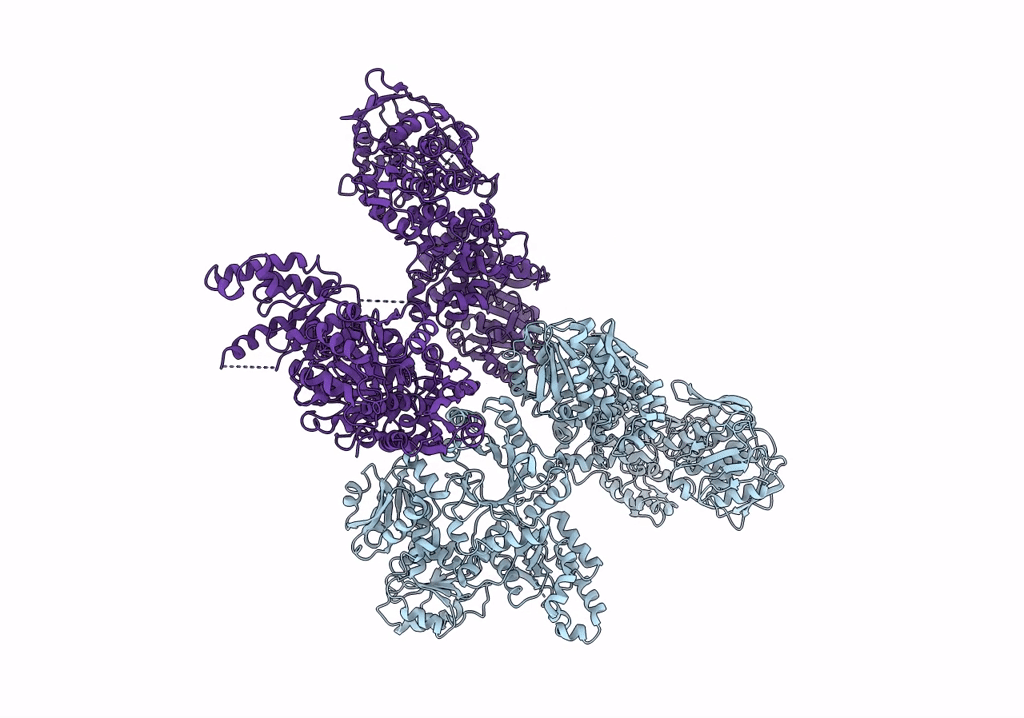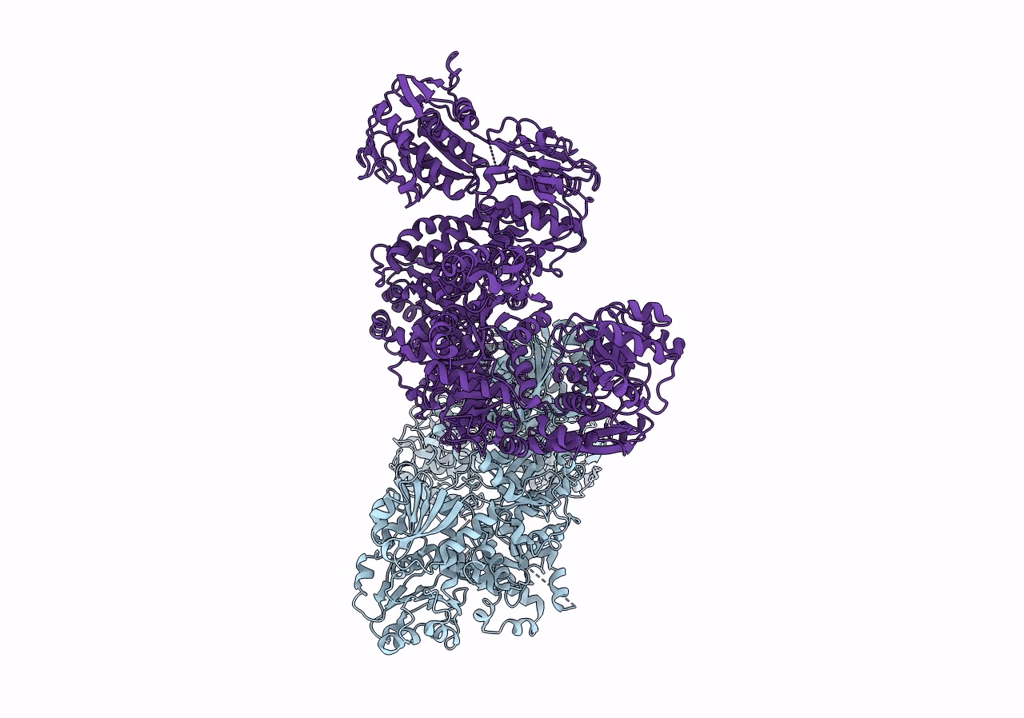
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Resolution:3.46 Å Release Date: 2022-11-30 Classification: PROTON TRANSPORT Ligands: HEM, HEO, CU, 3PE |
 |
Organism: Candida albicans
Method: ELECTRON MICROSCOPY Release Date: 2022-05-04 Classification: BIOSYNTHETIC PROTEIN |
 |
Organism: Candida albicans
Method: ELECTRON MICROSCOPY Release Date: 2022-05-04 Classification: BIOSYNTHETIC PROTEIN |
 |
Organism: Candida albicans
Method: ELECTRON MICROSCOPY Release Date: 2022-05-04 Classification: BIOSYNTHETIC PROTEIN |
 |
Crystal Structure Of The 5-Enolpyruvate-Shikimate-3-Phosphate Synthase (Epsps) Domain Of Aro1 From Candida Albicans In Complex With Shikimate-3-Phosphate
Organism: Candida albicans ca6
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2022-03-16 Classification: TRANSFERASE Ligands: TRS, S3P |
 |
Crystal Structure Of The Shikimate Kinase + 3-Dehydroquinate Dehydratase + 3-Dehydroshikimate Dehydrogenase Domains Of Aro1 From Candida Albicans
Organism: Candida albicans ca6
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2022-03-16 Classification: TRANSFERASE,OXIDOREDUCTASE Ligands: MG, CL, GOL |
 |
Structure Of Human Respiratory Syncytial Virus Nonstructural Protein 2 (Ns2)
Organism: Human respiratory syncytial virus a
Method: X-RAY DIFFRACTION Resolution:2.82 Å Release Date: 2021-03-17 Classification: VIRAL PROTEIN Ligands: TAR, CL |
 |
Single Particle Reconstruction Of Glucose Isomerase From Streptomyces Rubiginosus Based On Data Acquired In The Presence Of Substantial Aberrations
Organism: Streptomyces rubiginosus
Method: ELECTRON MICROSCOPY Resolution:2.70 Å Release Date: 2020-02-19 Classification: ISOMERASE Ligands: MN |
 |
Single Particle Reconstruction Of Hemq From Geobacillus Based On Data Acquired In The Presence Of Substantial Aberrations
Organism: Geobacillus sp. (strain y412mc52)
Method: ELECTRON MICROSCOPY Resolution:2.32 Å Release Date: 2020-02-19 Classification: OXIDOREDUCTASE |
 |
Single Particle Reconstruction Of Hemq From Geobacillus Based On Data Acquired In The Presence Of Substantial Aberrations
Organism: Geobacillus sp. (strain y412mc52)
Method: ELECTRON MICROSCOPY Resolution:2.60 Å Release Date: 2020-02-19 Classification: OXIDOREDUCTASE |
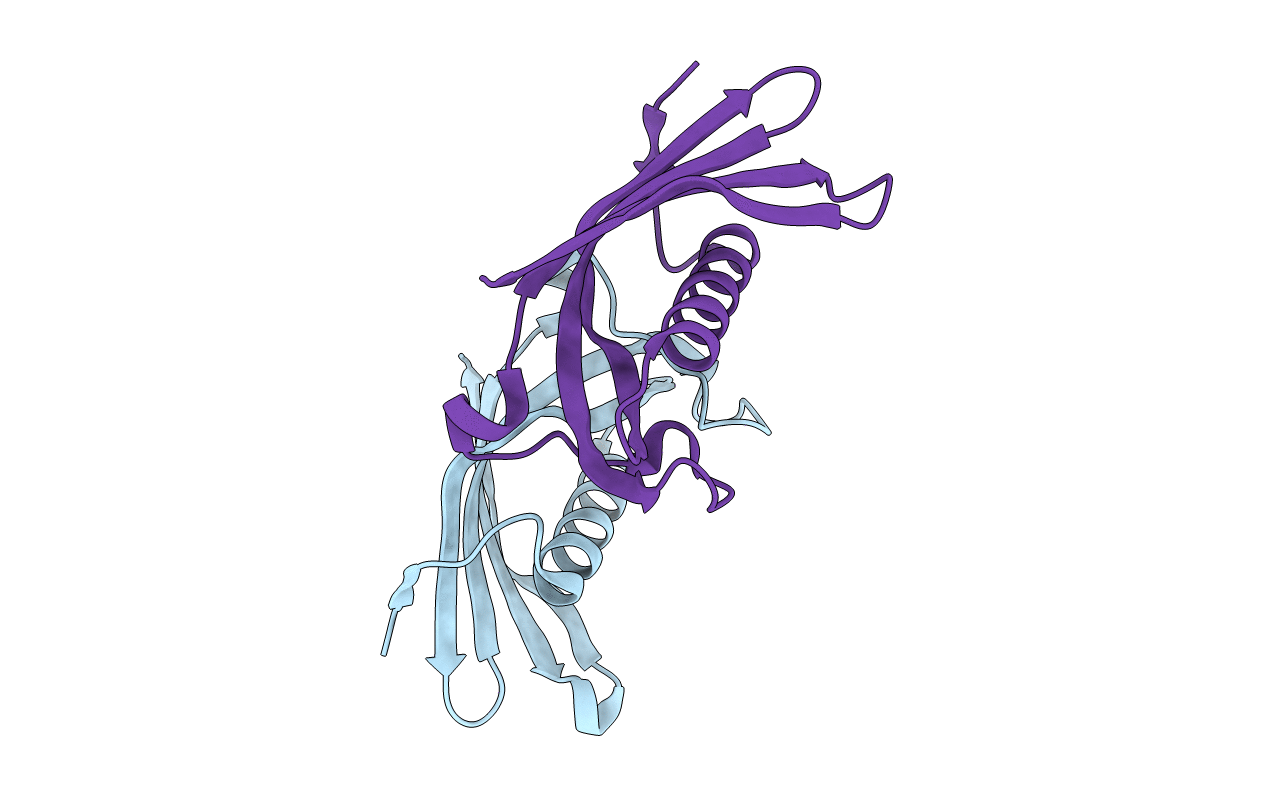 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.65 Å Release Date: 2019-08-07 Classification: HYDROLASE |
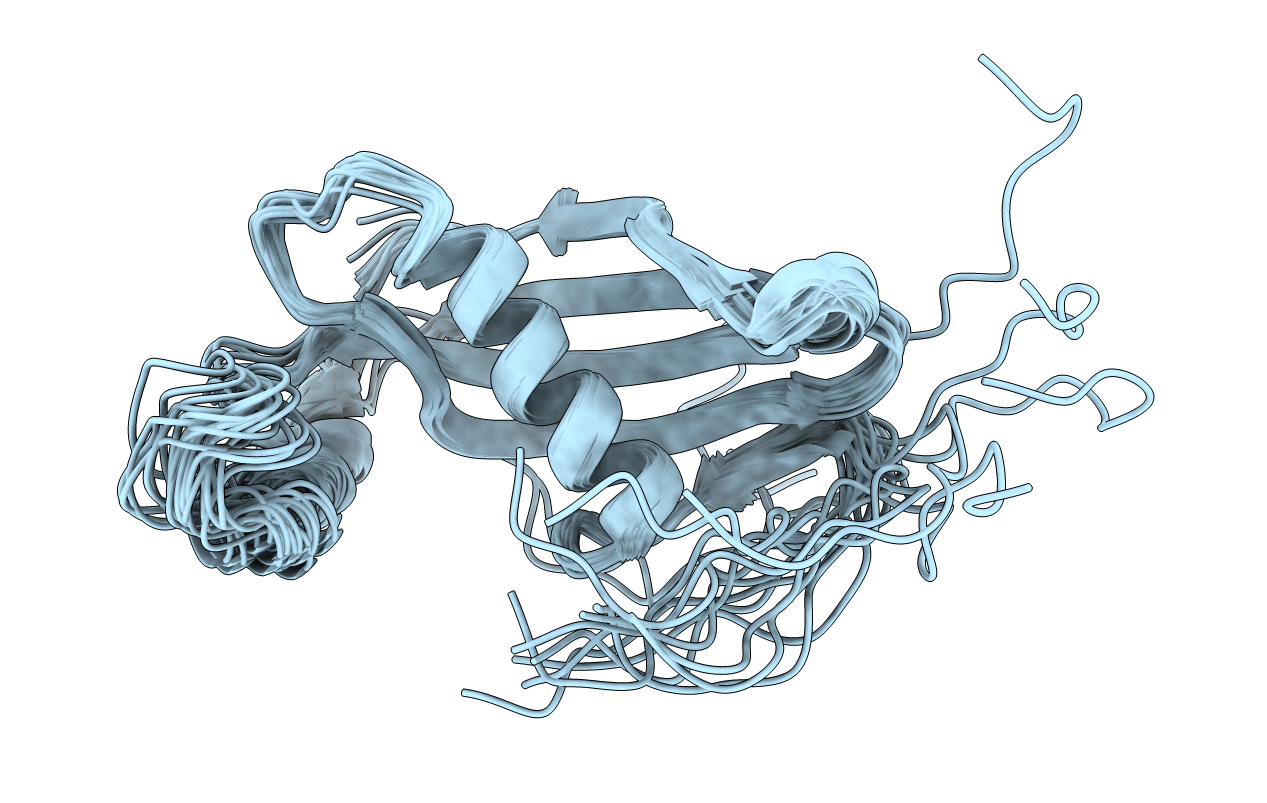 |
Extremely Stable Monomeric Variant Of Human Cystatin C With Single Amino Acid Substitution
Organism: Homo sapiens
Method: SOLUTION NMR, SOLUTION SCATTERING Release Date: 2019-07-31 Classification: HYDROLASE |
 |
Organism: Aspergillus terreus
Method: X-RAY DIFFRACTION Resolution:1.38 Å Release Date: 2018-11-14 Classification: HYDROLASE Ligands: NAG, CA, 1PE, PG4, PGE, PG0, EDO, DTU, ACT, MPO, BGC |
 |
Organism: Streptomyces rubiginosus
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2018-05-02 Classification: ISOMERASE,Oxidoreductase Ligands: MRD, MN, CL, CA |
 |
Crystal Structure Of The 3-Dehydroquinate Synthase (Dhqs) Domain Of Aro1 From Candida Albicans Sc5314 In Complex With Nadh
Organism: Candida albicans (strain sc5314 / atcc mya-2876)
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2018-01-24 Classification: LYASE Ligands: NAD, EDO, CL |
 |
Crystal Structure Of Native Beta-N-Acetylhexosaminidase Isolated From Aspergillus Oryzae
Organism: Aspergillus oryzae
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2017-12-27 Classification: HYDROLASE Ligands: MAN, NAG, NGT, CL, TBR |

