Search Count: 237
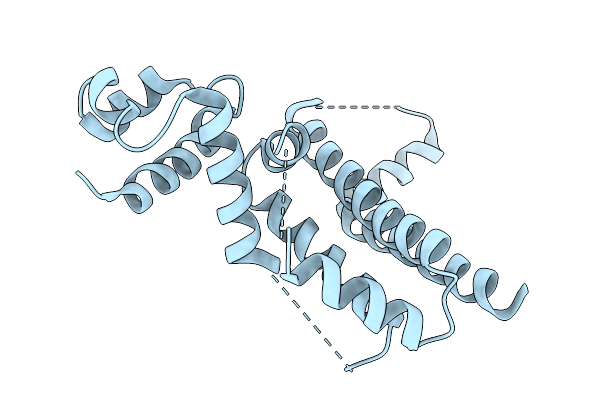 |
Crystal Structure Of The Engineered Sulfonylurea Repressor Esr (L7-D1), Apo Form
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Release Date: 2025-10-08 Classification: DNA BINDING PROTEIN |
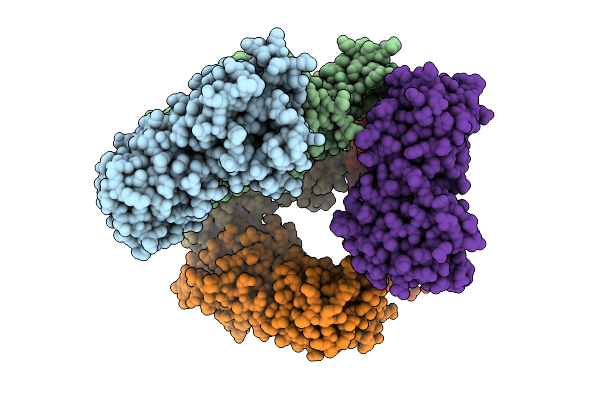 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Release Date: 2025-07-16 Classification: DNA BINDING PROTEIN Ligands: RIP |
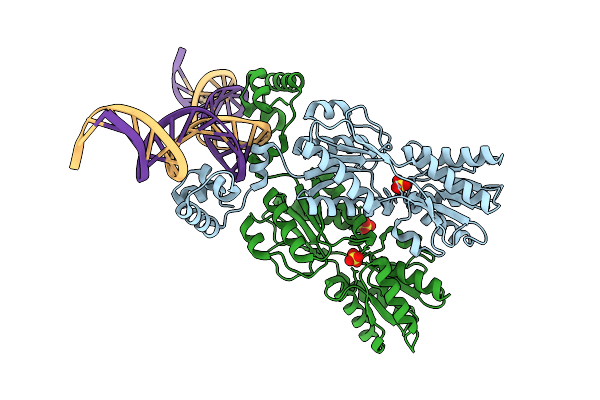 |
Crystal Structure Of The Ribose Operon Repressor, Rbsr, Bound To Ribose Operon
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Release Date: 2025-07-16 Classification: DNA BINDING PROTEIN/DNA Ligands: SO4 |
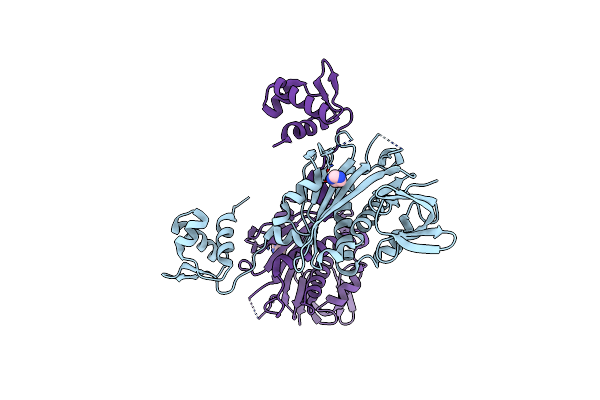 |
Bifunctional Ligase/Repressor Bira From Klebsiella Pneumoniae (Domain Swapped Dimer)
Organism: Klebsiella pneumoniae subsp. pneumoniae 1158
Method: X-RAY DIFFRACTION Resolution:2.85 Å Release Date: 2022-11-30 Classification: TRANSCRIPTION Ligands: IMD |
 |
Organism: Escherichia coli k-12
Method: X-RAY DIFFRACTION Resolution:2.25 Å Release Date: 2022-06-01 Classification: TRANSCRIPTION Ligands: URA, EDO |
 |
The Crystal Structure Of Escherichia Coli Murr In Complex With N-Acetylglucosamine-6-Phosphate
Organism: Escherichia coli k-12
Method: X-RAY DIFFRACTION Resolution:1.25 Å Release Date: 2022-04-20 Classification: GENE REGULATION Ligands: 4QY, GOL, MXE |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.28 Å Release Date: 2022-04-20 Classification: GENE REGULATION Ligands: PO4 |
 |
The Crystal Structure Of Escherichia Coli Murr In Complex With N-Acetylmuramic-Acid-6-Phosphate
Organism: Escherichia coli k-12
Method: X-RAY DIFFRACTION Resolution:1.22 Å Release Date: 2022-04-20 Classification: GENE REGULATION Ligands: J79 |
 |
Organism: Bacillus subtilis
Method: X-RAY DIFFRACTION Resolution:2.45 Å Release Date: 2021-12-22 Classification: DNA BINDING PROTEIN Ligands: G4P |
 |
Crystal Structure Of The Dna-Binding Transcriptional Repressor Deor From Escherichia Coli Str. K-12
Organism: Escherichia coli (strain k12)
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2021-12-01 Classification: DNA BINDING PROTEIN Ligands: EDO, SO4, CL |
 |
Organism: Pseudomonas putida
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2021-10-06 Classification: TRANSCRIPTION Ligands: MG |
 |
Organism: Pseudomonas putida
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2021-10-06 Classification: TRANSCRIPTION Ligands: STL, MG |
 |
Organism: Pseudomonas putida
Method: X-RAY DIFFRACTION Resolution:1.71 Å Release Date: 2021-10-06 Classification: TRANSCRIPTION Ligands: STL, MG |
 |
Crystal Structure Of Domain Swapped Trp Repressor V58I Variant With Purification Tag
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.45 Å Release Date: 2021-07-14 Classification: DNA BINDING PROTEIN Ligands: IMD |
 |
Organism: Escherichia coli (strain k12)
Method: X-RAY DIFFRACTION Resolution:2.05 Å Release Date: 2020-09-30 Classification: DNA BINDING PROTEIN Ligands: IPA |
 |
Crystal Structure Of Domain Swapped Trp Repressor V58I Variant With Bound L-Trp
Organism: Escherichia coli (strain k12)
Method: X-RAY DIFFRACTION Resolution:2.45 Å Release Date: 2020-09-30 Classification: DNA BINDING PROTEIN Ligands: TRP, IPA |
 |
Organism: Escherichia coli k-12
Method: X-RAY DIFFRACTION Resolution:3.50 Å Release Date: 2019-10-09 Classification: GENE REGULATION |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.13 Å Release Date: 2019-03-27 Classification: TRANSCRIPTION Ligands: IOP |
 |
Tryptophan Repressor Trpr From E.Coli Wildtype With Indole-3-Acetic Acid As Ligand
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Release Date: 2019-02-06 Classification: TRANSCRIPTION Ligands: IAC |
 |
Tryptophan Repressor Trpr From E.Coli Variant S88Y With Indole-3-Acetic Acid As Ligand
Organism: Escherichia coli (strain 55989 / eaec)
Method: X-RAY DIFFRACTION Release Date: 2019-02-06 Classification: TRANSCRIPTION Ligands: IAC, EDO, SO4 |

