Search Count: 59
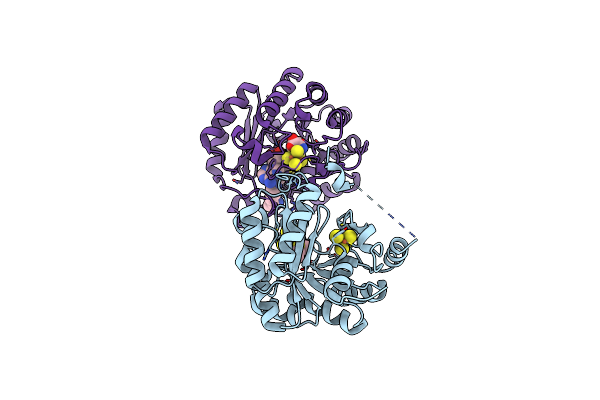 |
Structure Of Human Lias In The Presence Of 5'-Deoxyadenosine And Octanoyl-Modified Peptide
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-06-11 Classification: BIOSYNTHETIC PROTEIN Ligands: SF4, MET, 5AD, LYS, OCA |
 |
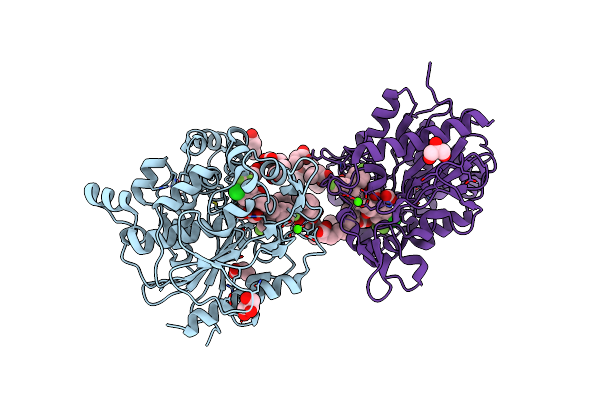 |
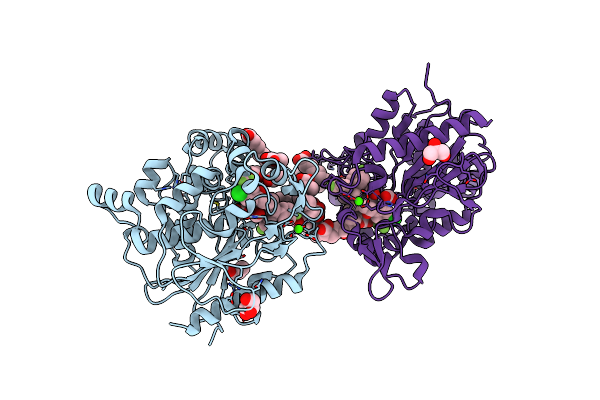
Organism: Staphylococcus aureus
Method: X-RAY DIFFRACTION Release Date: 2025-04-30 Classification: HYDROLASE Ligands: OCA, PPI, GOL, A1L60, FMT, ZN, CA, 6NA, CL, BUA, 11A |
 |
Single Particle Cryo-Em Reconstruction Of Oryza Sativa Recombinant Human Serum Albumin
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-03-19 Classification: PROTEIN BINDING Ligands: OCA |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-03-19 Classification: TRANSPORT PROTEIN Ligands: OCA |
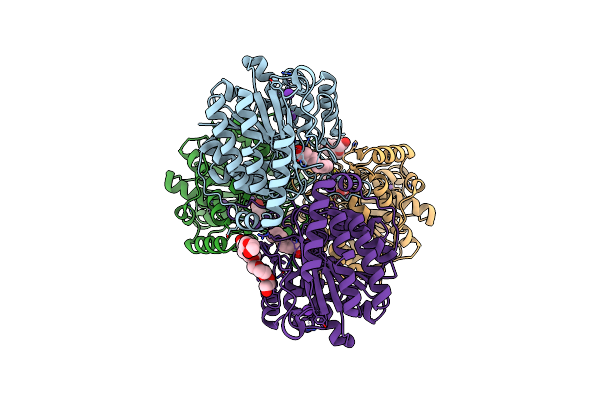 |
Crystal Structure Of Dimethoate Hydrolase (Dmha) Of Rhizorhabdus Wittichii In Complex With Octanoic Acid
Organism: Rhizorhabdus wittichii dc-6
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2024-11-06 Classification: HYDROLASE Ligands: OCA, K, ZN, 1PE |
 |
Crystal Structure Of Rhizorhabdus Wittichii Dimethoate Hydrolase (Dmha) In Complex With Saha
Organism: Rhizorhabdus wittichii dc-6
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2024-11-06 Classification: HYDROLASE Ligands: SHH, ZN, K, OCA |
 |
Crystal Structure Of Beta-Ketoacyl-Acp Synthase Fabf K336A In Complex With Octanoyl-Acp From Helicobacter Pylori
Organism: Helicobacter pylori
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2024-10-09 Classification: BIOSYNTHETIC PROTEIN Ligands: OCA, PN7 |
 |
 |
 |
 |
The Structure Of Thermomyces Lanuginosa Lipase In Complex With 1,3 Diacylglycerol In A Monoclinic Crystal Form
Organism: Thermomyces lanuginosus
Method: X-RAY DIFFRACTION Resolution:2.48 Å Release Date: 2024-05-01 Classification: HYDROLASE Ligands: OCA, CA, NAG, LTV, PO4 |
 |
High Resolution Structure Of The Monoclinic Form Of Thermomyces Lanuginosa Lipase Complexed With Its Catalytic Products
Organism: Thermomyces lanuginosus
Method: X-RAY DIFFRACTION Resolution:1.43 Å Release Date: 2024-05-01 Classification: HYDROLASE Ligands: OCA, NAG, LTV, PO4, CA |
 |
Cryo-Em Structure Of The Photosystem I - Lhci Supercomplex From Coelastrella Sp.
|
 |
Organism: Streptomyces nodosus
Method: X-RAY DIFFRACTION Resolution:1.43 Å Release Date: 2024-02-07 Classification: OXIDOREDUCTASE Ligands: HEM, OCA |
 |
Human Consensus Olfactory Receptor Or52C In Complex With Octanoic Acid (Oca) And G Protein
Organism: Homo sapiens, Lama glama
Method: ELECTRON MICROSCOPY Release Date: 2023-12-20 Classification: MEMBRANE PROTEIN Ligands: OCA |
 |
Crystal Structure Of Cyp109A2 From Bacillus Megaterium Bound With Putative Ligands Hexanoic Acid And Octanoic Acid
Organism: Priestia megaterium dsm 319
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2023-07-12 Classification: OXIDOREDUCTASE Ligands: HEB, 6NA, OCA, SO4 |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2023-03-01 Classification: TRANSPORT PROTEIN Ligands: UZC, DMS, MYR, GOL, OCA, SO4 |
 |
Crystal Structure Of Sars Cov-2 Main Protease In Complex With An Inhibitor 58
Organism: Severe acute respiratory syndrome coronavirus 2, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.94 Å Release Date: 2022-12-07 Classification: ANTIVIRAL PROTEIN Ligands: DMS, OCA |
 |
Organism: Alcanivorax dieselolei
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2022-11-02 Classification: OXIDOREDUCTASE Ligands: HEM, OCA |

