Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
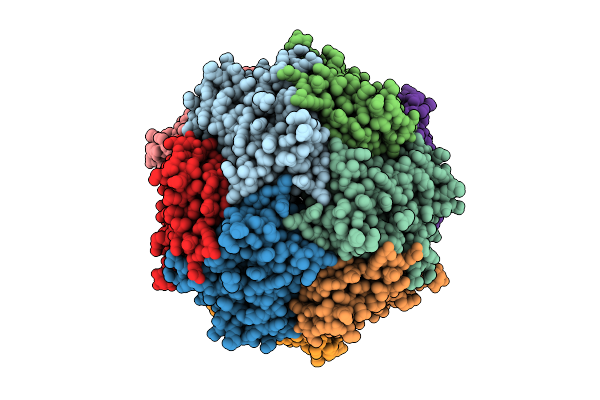 |
Structure Of Thioferritin (Pfdpsl) With Ferrihydrite Growth At A Single Three-Fold Pore.
Organism: Pyrococcus furiosus
Method: ELECTRON MICROSCOPY Release Date: 2025-10-29 Classification: METAL BINDING PROTEIN Ligands: FE, OXY, O |
 |
Organism: Physeter catodon
Method: X-RAY DIFFRACTION Release Date: 2025-10-15 Classification: OXIDOREDUCTASE Ligands: HEM, O |
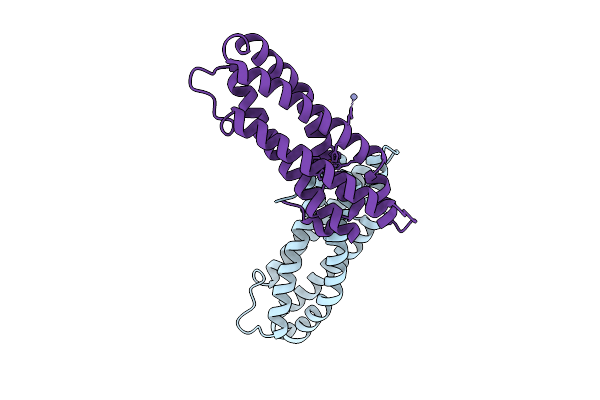 |
Crystal Structure Of Staphylococcus Aureus Cysteine-Free Scda With Bound Iron, Determined By Molecular Replacement And Fe Anomalous Signal
Organism: Staphylococcus aureus
Method: X-RAY DIFFRACTION Release Date: 2025-09-17 Classification: METAL BINDING PROTEIN Ligands: FE, O, ZN |
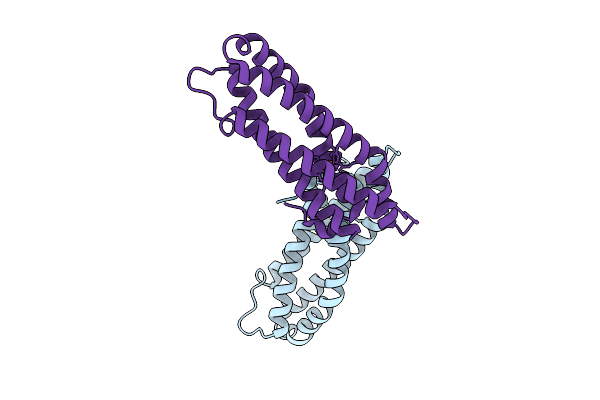 |
Organism: Staphylococcus aureus
Method: X-RAY DIFFRACTION Release Date: 2025-08-13 Classification: OXYGEN BINDING Ligands: FE, O |
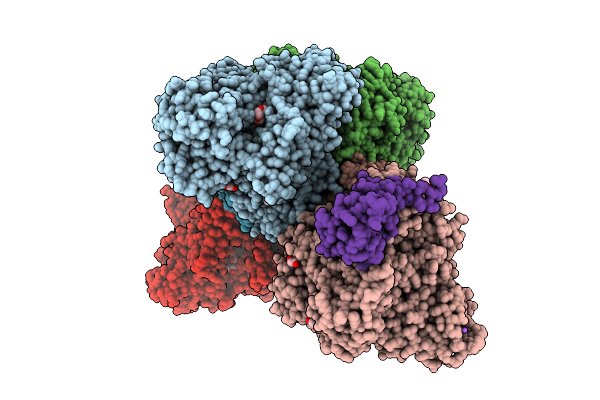 |
Organism: Alcaligenes faecalis
Method: X-RAY DIFFRACTION Release Date: 2025-08-06 Classification: OXIDOREDUCTASE Ligands: MGD, 4MO, F3S, PEG, EDO, 1PE, PGE, GOL, O, NA, FES, P4G, PG0 |
 |
Organism: Alcaligenes faecalis
Method: X-RAY DIFFRACTION Release Date: 2025-08-06 Classification: OXIDOREDUCTASE Ligands: MO, MGD, F3S, EDO, PEG, SBO, P33, PGE, O, NA, FES, GOL, 4MO, P4G, 1PE, PG0 |
 |
Structure Of Thioferritin Exhibiting Iron Mineral Nucleation, From Pyrococcus Furiosis
Organism: Pyrococcus furiosus
Method: ELECTRON MICROSCOPY Release Date: 2025-08-06 Classification: METAL BINDING PROTEIN Ligands: FE, O |
 |
Organism: Staphylococcus aureus
Method: X-RAY DIFFRACTION Release Date: 2025-07-09 Classification: METAL BINDING PROTEIN Ligands: FE, O, ZN |
 |
Organism: Clostridioides difficile p28
Method: X-RAY DIFFRACTION Release Date: 2025-06-11 Classification: OXIDOREDUCTASE Ligands: FE, O |
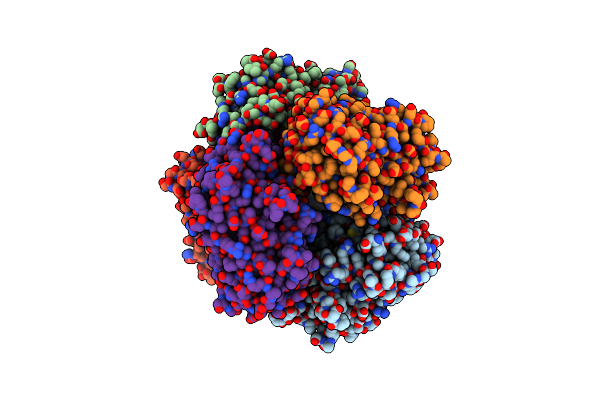 |
Organism: Streptomyces lividans 1326
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2025-05-28 Classification: OXIDOREDUCTASE Ligands: HEM, MG, O |
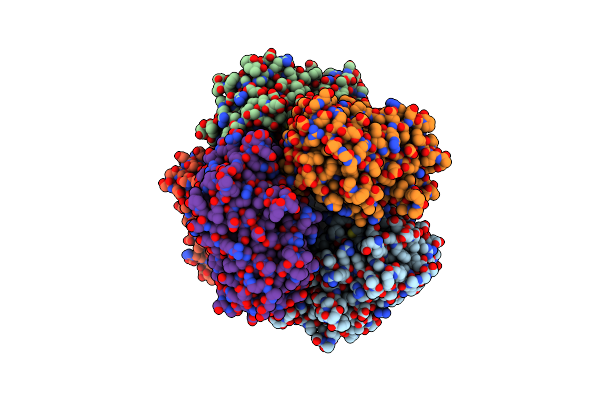 |
Organism: Streptomyces lividans 1326
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2025-05-28 Classification: OXIDOREDUCTASE Ligands: HEM, MG, O |
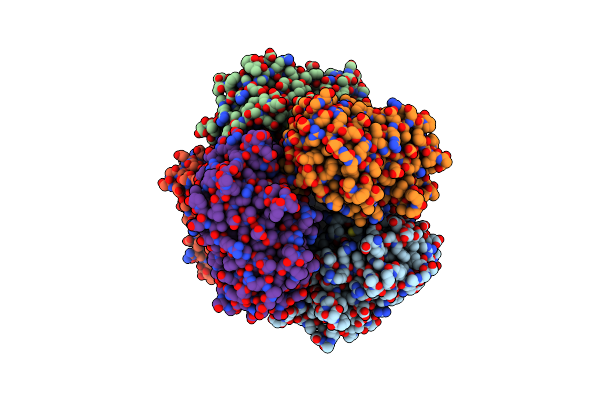 |
Organism: Streptomyces lividans 1326
Method: X-RAY DIFFRACTION Resolution:1.96 Å Release Date: 2025-05-28 Classification: OXIDOREDUCTASE Ligands: HEM, MG, O |
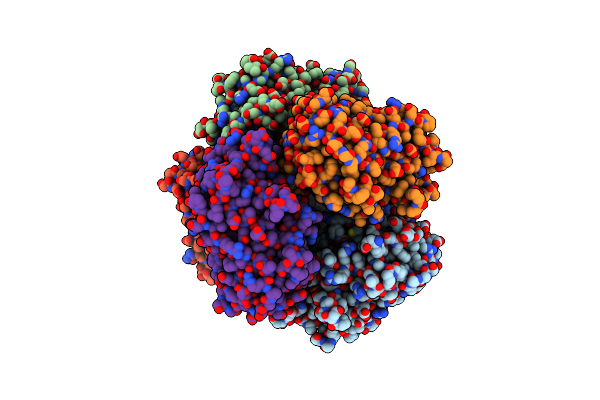 |
Organism: Streptomyces lividans 1326
Method: X-RAY DIFFRACTION Resolution:1.96 Å Release Date: 2025-05-28 Classification: OXIDOREDUCTASE Ligands: HEM, O, MG |
 |
Organism: Streptomyces lividans 1326
Method: X-RAY DIFFRACTION Resolution:1.96 Å Release Date: 2025-05-28 Classification: OXIDOREDUCTASE Ligands: HEM, O, MG |
 |
Organism: Pseudorhizobium banfieldiae
Method: X-RAY DIFFRACTION Resolution:1.89 Å Release Date: 2025-05-21 Classification: OXIDOREDUCTASE Ligands: SER, 4MO, F3S, EDO, PEG, MGD, SO4, P33, GOL, O, FES, 1PE |
 |
Organism: Streptomyces lividans 1326
Method: X-RAY DIFFRACTION Resolution:1.83 Å Release Date: 2025-04-16 Classification: OXIDOREDUCTASE Ligands: HEM, MG, O, PG4 |
 |
Organism: Streptomyces lividans 1326
Method: X-RAY DIFFRACTION Resolution:1.92 Å Release Date: 2025-04-16 Classification: OXIDOREDUCTASE Ligands: HEM, MG, O, PG4 |
 |
Crystal Structure Of The Functional Unit (Hth1-H) Of Hemocyanin From Haliotis Discus Hannai,Containing A Met Site
Organism: Haliotis discus hannai
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2025-01-15 Classification: METAL BINDING PROTEIN Ligands: MAN, EOH, CU, OH, O |
 |
Organism: Pseudonocardia thermophila
Method: X-RAY DIFFRACTION Resolution:1.29 Å Release Date: 2024-09-25 Classification: LYASE Ligands: O |
 |
Organism: Streptomyces lividans 1326
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2024-08-07 Classification: OXIDOREDUCTASE Ligands: O, HEM, MG, PG4 |