Search Count: 14
 |
Organism: Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2023-10-11 Classification: TRANSPORT PROTEIN |
 |
Organism: Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2023-10-11 Classification: TRANSPORT PROTEIN |
 |
Organism: Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2023-10-11 Classification: TRANSPORT PROTEIN |
 |
Crystal Structure Of Hsting In Complex With C[2',3'-(Ribo-2'-G, Xylo-3'-A)-Mp](Rj244)
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2022-10-19 Classification: IMMUNE SYSTEM Ligands: 9UR |
 |
Crystal Structure Of Hsting In Complex With C[2',3'-(Ara-2'-G, Ribo-3'-A)-Mp] (Rj242)
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.25 Å Release Date: 2022-10-12 Classification: IMMUNE SYSTEM Ligands: 9UH |
 |
Organism: Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2022-01-26 Classification: TRANSLOCASE |
 |
Organism: Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2022-01-26 Classification: TRANSLOCASE |
 |
Organism: Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2022-01-26 Classification: TRANSLOCASE |
 |
Structure Of The 44 Kda Complex Of Interferon-Alpha2 With The Extracellular Part Of Ifnar2 Obtained By 2D-Double Difference Noesy
|
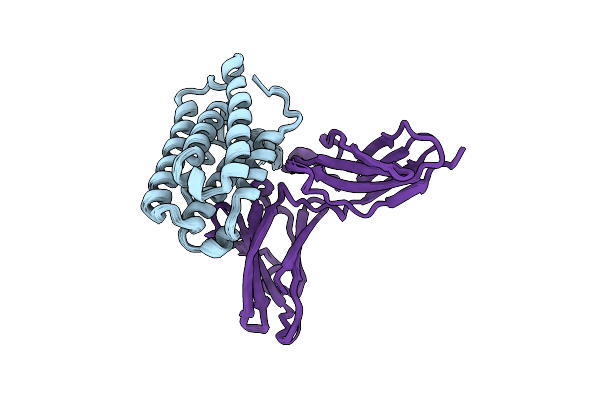 |
Inter-Molecular Interactions In A 44 Kda Interferon-Receptor Complex Detected By Asymmetric Back-Protonation And 2D Noesy
Organism: Homo sapiens
Method: SOLUTION NMR Release Date: 2010-06-23 Classification: ANTIVIRAL PROTEIN |
 |
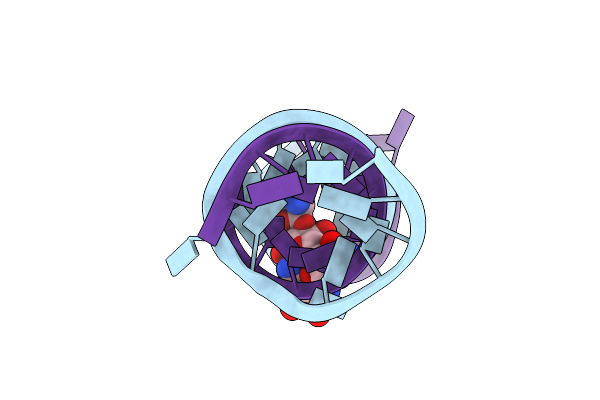
Crystal Structure Of The Homo Sapiens Cytoplasmic Ribosomal Decoding Site Complexed With Paromamine Derivative Nb33
Method: X-RAY DIFFRACTION
Resolution:2.80 Å Release Date: 2007-11-06 Classification: RNA Ligands: N33 |
 |
Crystal Structure Of The Homo Sapiens Cytoplasmic Ribosomal Decoding Site In Presence Of Paromomycin
Method: X-RAY DIFFRACTION
Resolution:2.80 Å Release Date: 2007-11-06 Classification: RNA Ligands: PAR |
 |
Crystal Structure Of The Prokaryotic Ribosomal Decoding Site Complexed With Paromamine Derivative Nb30
Method: X-RAY DIFFRACTION
Resolution:2.90 Å Release Date: 2007-11-06 Classification: RNA Ligands: N30 |
 |
Crystal Structure Of The Homo Sapiens Cytoplasmic Ribosomal Decoding Site In Presence Of Paromamine Derivative Nb30
Method: X-RAY DIFFRACTION
Resolution:2.70 Å Release Date: 2007-11-06 Classification: RNA Ligands: SPM |

