Search Count: 19
 |
Organism: Rhodococcus wratislaviensis
Method: X-RAY DIFFRACTION Resolution:2.75 Å Release Date: 2021-01-06 Classification: OXIDOREDUCTASE Ligands: ZN, GOL, SO4, FE, CL |
 |
Organism: Rhodococcus wratislaviensis
Method: X-RAY DIFFRACTION Resolution:2.45 Å Release Date: 2021-01-06 Classification: OXIDOREDUCTASE Ligands: ZN, SO4, EDO, FE |
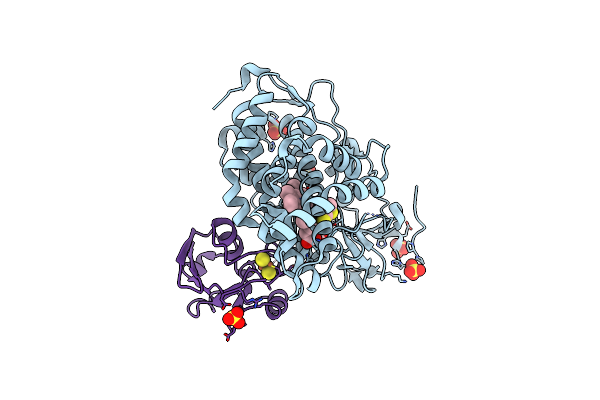 |
High-Resolution Crystal Structure Of The Electron Transfer Complex Of Cytochrome P450Cam With Putidaredoxin
Organism: Pseudomonas putida
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2017-01-18 Classification: OXIDOREDUCTASE/ELECTRON TRANSPORT Ligands: HEM, SO4, DTT, FES |
 |
Crystal Structure Of The Electron-Transfer Complex Of Copper Nitrite Reductase With A Cupredoxin
Organism: Alcaligenes xylosoxydans xylosoxydans, Hyphomicrobium denitrificans
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2016-11-23 Classification: OXIDOREDUCTASE/ELECTRON TRANSPORT Ligands: CU |
 |
Crystal Structure Of The Chloride-Bound Form Of Blue Copper Nitrite Reductase
Organism: Alcaligenes xylosoxydans xylosoxydans
Method: X-RAY DIFFRACTION Resolution:1.35 Å Release Date: 2016-11-23 Classification: OXIDOREDUCTASE Ligands: CU, CL, PG4 |
 |
Copper-Containing Nitrite Reductase From Thermophilic Bacterium Geobacillus Thermodenitrificans (Re-Refined)
Organism: Geobacillus thermodenitrificans ng80-2
Method: X-RAY DIFFRACTION Resolution:1.15 Å Release Date: 2015-05-20 Classification: OXIDOREDUCTASE Ligands: CU, CL, MPD, PEG, EDO, SO4, ACY, TRS |
 |
Crystal Structure Of Copper Nitrite Reductase From Geobacillus Kaustophilus
Organism: Geobacillus kaustophilus
Method: X-RAY DIFFRACTION Resolution:1.30 Å Release Date: 2014-07-23 Classification: OXIDOREDUCTASE Ligands: CU, ZN, NA, ACY, MPD, SO4 |
 |
Crystal Structure Of The N-Terminal 1-37 Residues Deleted Mutant Of Geobacillus Copper Nitrite Reductase
Organism: Geobacillus kaustophilus
Method: X-RAY DIFFRACTION Resolution:1.77 Å Release Date: 2014-07-23 Classification: OXIDOREDUCTASE Ligands: CU, FMT |
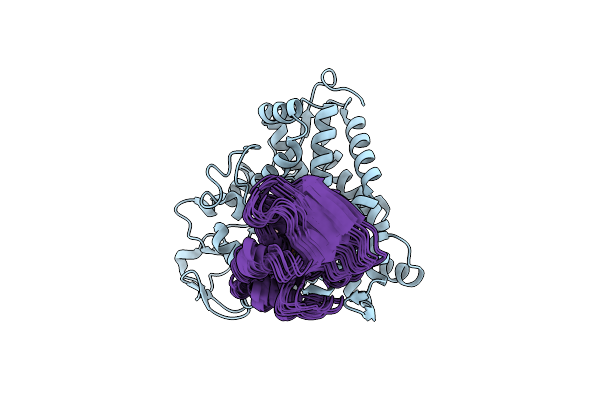 |
The Structure Of The Complex Of Cytochrome P450Cam And Its Electron Donor Putidaredoxin Determined By Paramagnetic Nmr Spectroscopy
Organism: Pseudomonas putida
Method: SOLUTION NMR Release Date: 2013-08-21 Classification: OXIDOREDUCTASE/METAL BINDING PROTEIN Ligands: HEM, CAM, FES |
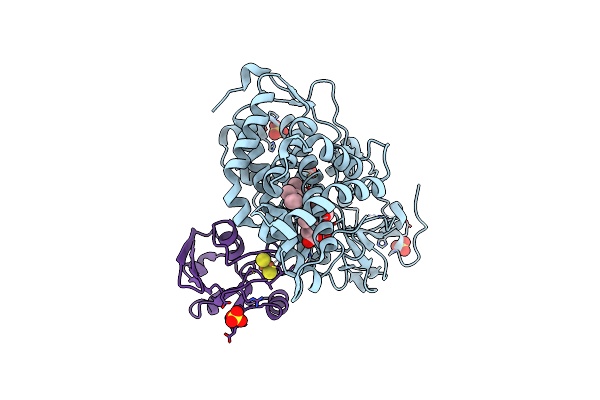 |
Crystal Structure Of The Electron Transfer Complex Of Cytochrome P450Cam With Putidaredoxin
Organism: Pseudomonas putida
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2013-08-21 Classification: OXIDOREDUCTASE/ELECTRON TRANSPORT Ligands: HEM, SO4, GOL, FES |
 |
Crystal Structure Of Electron Transfer Complex Of Nitrite Reductase With Cytochrome C
Organism: Achromobacter xylosoxidans
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2009-06-09 Classification: OXIDOREDUCTASE/electron transport Ligands: CU, HEM |
 |
Crystal Structure Of Nitrite Reductase From Pseudoalteromonas Haloplanktis Tac125
Organism: Pseudoalteromonas haloplanktis
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2009-06-09 Classification: OXIDOREDUCTASE Ligands: CU, HEM, SO4 |
 |
Organism: Hyphomicrobium denitrificans
Method: X-RAY DIFFRACTION Resolution:1.18 Å Release Date: 2008-12-30 Classification: ELECTRON TRANSPORT Ligands: CU, PO4 |
 |
Organism: Hyphomicrobium denitrificans
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2007-02-20 Classification: OXIDOREDUCTASE Ligands: CU, K |
 |
Organism: Hyphomicrobium denitrificans
Method: X-RAY DIFFRACTION Resolution:1.98 Å Release Date: 2006-08-10 Classification: ELECTRON TRANSPORT Ligands: ZN, HEM |
 |
Crystal Structure Of Methanol Dehydrogenase From Hyphomicrobium Denitrificans
Organism: Hyphomicrobium denitrificans
Method: X-RAY DIFFRACTION Resolution:2.49 Å Release Date: 2006-08-09 Classification: OXIDOREDUCTASE Ligands: CA, PQQ |
 |
Complex Of Fe-Type Nhase With Cyclohexyl Isocyanide, Photo-Activated For 1Hr At 277K
Organism: Rhodococcus erythropolis
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2006-02-05 Classification: LYASE Ligands: FE, MG, CYI |
 |
Organism: Rhodococcus erythropolis
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2006-01-11 Classification: LYASE Ligands: FE, MG, NO, CYI |
 |
Organism: Rhodococcus erythropolis
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2006-01-11 Classification: LYASE Ligands: FE, NO, MG |

