Search Count: 29
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-07-30 Classification: NUCLEAR PROTEIN Ligands: A1MAM, EDO, ACT |
 |
Organism: Streptomyces sp.
Method: X-RAY DIFFRACTION Resolution:2.12 Å Release Date: 2023-11-01 Classification: BIOSYNTHETIC PROTEIN Ligands: SAH, TRS |
 |
Crystal Structure Of Photosystem Ii From An Sqdg-Deficient Mutant Of Thermosynechococcus Elongatus
|
 |
 |
 |
 |
 |
Crystal Structure Of Nitrile Hydratase Mutant Br56K Complexed With Trimethylacetonitrile, Photo-Activated For 25 Min
Organism: Rhodococcus erythropolis
Method: X-RAY DIFFRACTION Resolution:1.18 Å Release Date: 2016-01-27 Classification: LYASE Ligands: FE, TAN, MG, CL |
 |
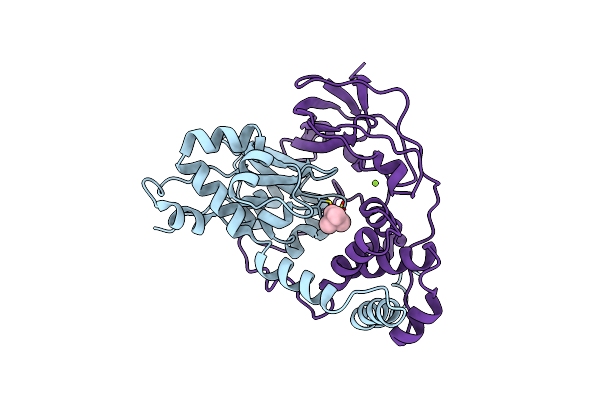
Crystal Structure Of Nitrile Hydratase Mutant Br56K Complexed With Trimethylacetonitrile, Photo-Activated For 120 Min
Organism: Rhodococcus erythropolis
Method: X-RAY DIFFRACTION Resolution:1.24 Å Release Date: 2016-01-27 Classification: LYASE Ligands: FE, MG, TAN |
 |
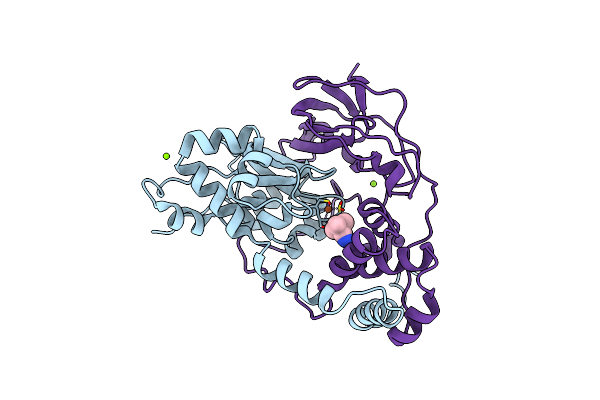
Crystal Structure Of Nitrile Hydratase Mutant Br56K Complexed With Trimethylacetonitrile, Photo-Activated For 700 Min
Organism: Rhodococcus erythropolis
Method: X-RAY DIFFRACTION Resolution:1.20 Å Release Date: 2016-01-27 Classification: LYASE Ligands: FE, TAN, CL, MG |
 |
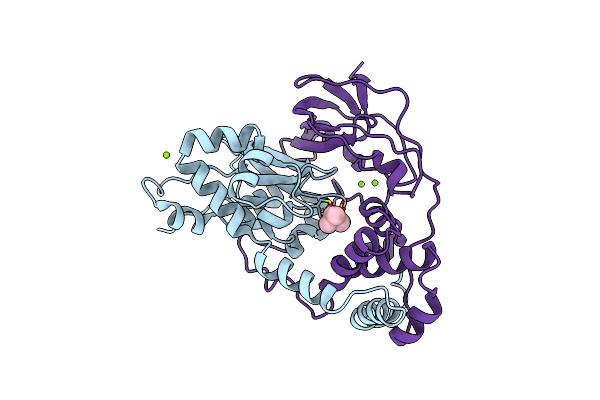
Crystal Structure Of Nitrile Hydratase Mutant Br56K Complexed With Trimethylacetonitrile, Photo-Activated For 5 Min
Organism: Rhodococcus erythropolis
Method: X-RAY DIFFRACTION Resolution:1.34 Å Release Date: 2016-01-27 Classification: LYASE Ligands: FE, MG, CL, TAN |
 |
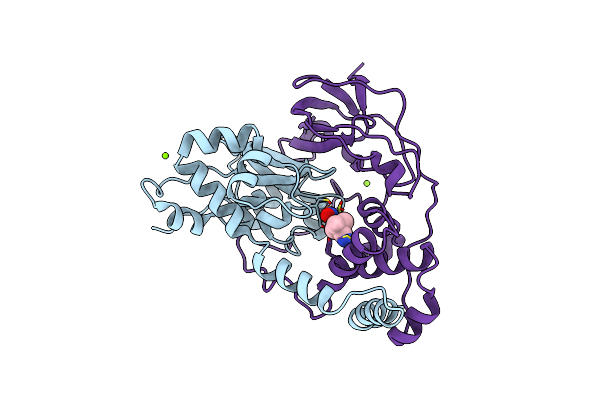
Crystal Structure Of Nitrile Hydratase Mutant Br56K Complexed With Trimethylacetonitrile, Photo-Activated For 50 Min
Organism: Rhodococcus erythropolis
Method: X-RAY DIFFRACTION Resolution:1.18 Å Release Date: 2015-06-17 Classification: LYASE Ligands: FE, MG, TAN |
 |
Crystal Structure Of Nitrile Hydratase Mutant Br56K Complexed With Trimethylacetonitrile, Before Photo-Activation
Organism: Rhodococcus erythropolis
Method: X-RAY DIFFRACTION Resolution:1.57 Å Release Date: 2015-06-17 Classification: LYASE Ligands: FE, NO, MG, CL, TAN |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2013-02-13 Classification: PROTEIN BINDING/METAL BINDING PROTEIN Ligands: ZN, K |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2013-02-13 Classification: PROTEIN BINDING/METAL BINDING PROTEIN Ligands: EDO, ZN, K |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.98 Å Release Date: 2013-02-13 Classification: PROTEIN BINDING/METAL BINDING PROTEIN Ligands: ZN, K |
 |
Crystal Structure Of Hcv Ns5B Rna Polymerase With A Novel Piperazine Inhibitor
Organism: Hepatitis c virus
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2012-08-08 Classification: HYDROLASE/HYDROLASE INHIBITOR Ligands: JT1, CL |
 |
Organism: Oryctolagus cuniculus
Method: ELECTRON MICROSCOPY Release Date: 2010-11-03 Classification: CONTRACTILE PROTEIN Ligands: ADP, PO4, MG |
 |
Crystal Structure Of A Dictyostelium P109A Mg2+-Actin In Complex With Human Gelsolin Segment 1
Organism: Homo sapiens, Dictyostelium discoideum
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2010-10-27 Classification: CONTRACTILE PROTEIN Ligands: CA, ADP, MG |
 |
Crystal Structure Of A Dictyostelium P109I Mg2+-Actin In Complex With Human Gelsolin Segment 1
Organism: Homo sapiens, Dictyostelium discoideum
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2010-10-27 Classification: CONTRACTILE PROTEIN Ligands: CA, MG, ATP |

