Search Count: 25
 |
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:2.47 Å Release Date: 2019-12-18 Classification: IMMUNE SYSTEM |
 |
Organism: Synthetic construct, Mus musculus
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2019-12-18 Classification: IMMUNE SYSTEM |
 |
Organism: Synthetic construct, Mus musculus
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2019-02-13 Classification: IMMUNE SYSTEM |
 |
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2019-02-13 Classification: IMMUNE SYSTEM |
 |
Organism: Bos taurus
Method: X-RAY DIFFRACTION Resolution:1.94 Å Release Date: 2012-05-23 Classification: HYDROLASE/HYDROLASE INHIBITOR Ligands: GHS, CA, E4B |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2011-02-23 Classification: CHAPERONE |
 |
Crystal Structure Of Human Hsp40 Hdj1 Peptide-Binding Domain Complexed With A C-Terminal Peptide Of Hsp70
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2011-02-23 Classification: CHAPERONE |
 |
Crystal Structure Of Human Hsp40 Hdj1 Peptide-Binding Domain Complexed With A C-Terminal Peptide Of Hsp70
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.51 Å Release Date: 2011-02-23 Classification: CHAPERONE |
 |
Crystal Structure Of The Sep22 Dodecamer, A Dps-Like Protein From Salmonella Enterica Subsp. Enterica Serovar Enteritidis
Organism: Salmonella enterica subsp. enterica serovar kentucky
Method: X-RAY DIFFRACTION Resolution:1.25 Å Release Date: 2011-01-12 Classification: METAL BINDING PROTEIN, Oxidoreductase Ligands: SO4, MG |
 |
Crystal Structure Of The Sep22 Dodecamer, A Dps-Like Protein From Salmonella Enterica Subsp. Enterica Serovar Enteritidis, Fe-Soaked Form
Organism: Salmonella enterica subsp. enterica serovar kentucky
Method: X-RAY DIFFRACTION Resolution:1.30 Å Release Date: 2011-01-12 Classification: METAL BINDING PROTEIN, Oxidoreductase Ligands: FE2, SO4, MG |
 |
Crystal Structure Of Ustilago Sphaerogena Ribonuclease U2 Complexed With Adenosine 3'-Monophosphate
Organism: Ustilago sphaerogena
Method: X-RAY DIFFRACTION Resolution:0.96 Å Release Date: 2010-07-07 Classification: HYDROLASE Ligands: 3AM, CA |
 |
Crystal Structure Of Ustilago Sphaerogena Ribonuclease U2 Complexed With Adenosine 3'-Monophosphate
Organism: Ustilago sphaerogena
Method: X-RAY DIFFRACTION Resolution:0.99 Å Release Date: 2010-07-07 Classification: HYDROLASE Ligands: 3AM, CA, CL |
 |
Organism: Ustilago sphaerogena
Method: X-RAY DIFFRACTION Resolution:1.32 Å Release Date: 2010-07-07 Classification: HYDROLASE Ligands: PO4, K, GOL |
 |
Crystal Structure Of Ustilago Sphaerogena Ribonuclease U2 Complexed With Adenosine 2'-Monophosphate
Organism: Ustilago sphaerogena
Method: X-RAY DIFFRACTION Resolution:1.03 Å Release Date: 2010-07-07 Classification: HYDROLASE Ligands: 2AM, CA |
 |
Solution Rna Structure Of Stem-Bulge-Stem Region Of The Hiv-1 Dimerization Initiation Site
|
 |
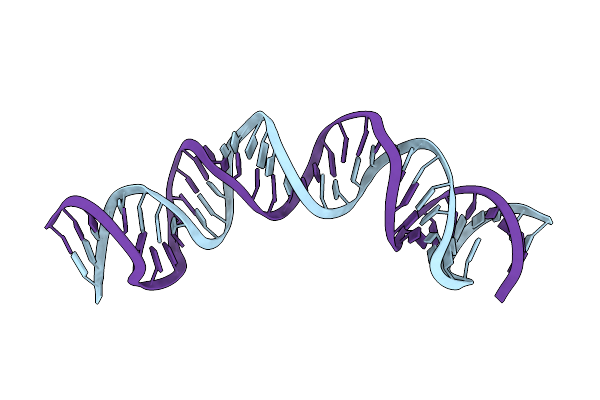
Solution Rna Structure Of Loop Region Of The Hiv-1 Dimerization Initiation Site In The Extended-Duplex Dimer
|
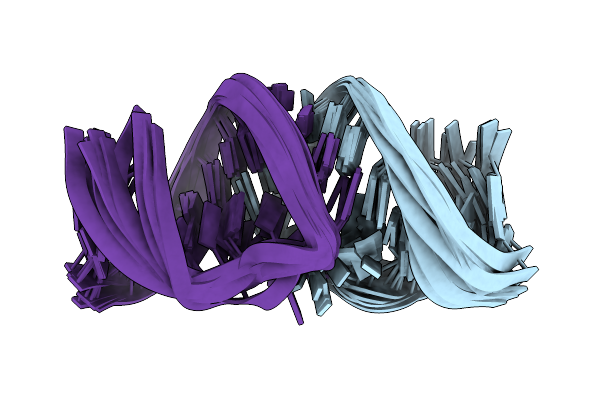 |
Solution Rna Structure Of Loop Region Of The Hiv-1 Dimerization Initiation Site In The Kissing-Loop Dimer
|
 |
Solution Rna Structure Model Of The Hiv-1 Dimerization Initiation Site In The Extended-Duplex Dimer
|
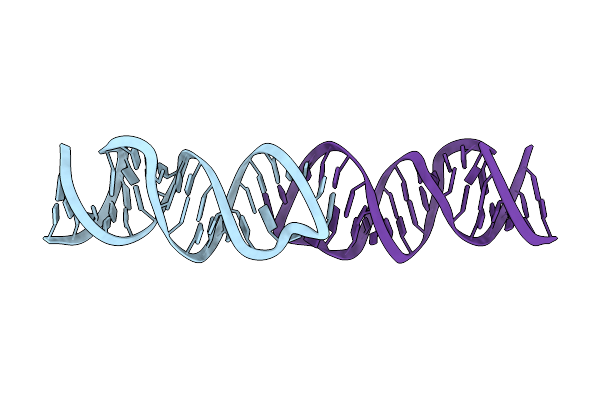 |
Solution Rna Structure Model Of The Hiv-1 Dimerization Initiation Site In The Kissing-Loop Dimer
|
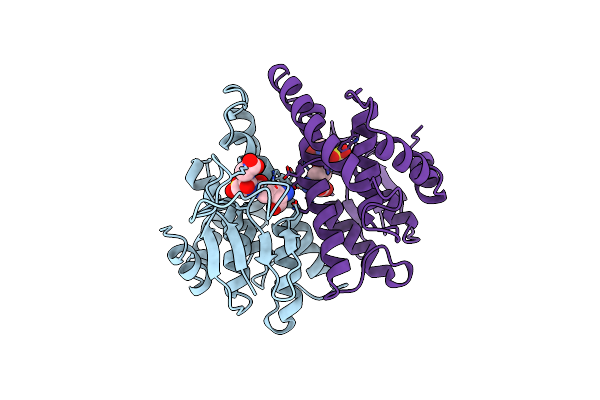 |
Crystal Structure Of Glutathione S-Transferase From Escherichia Coli Complexed With Glutathionesulfonic Acid
Organism: Escherichia coli k12
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 1999-01-13 Classification: TRANSFERASE Ligands: GTS |

