Search Count: 15
 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2024-03-27 Classification: RNA Ligands: K, DMS, W6F |
 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2024-03-27 Classification: RNA Ligands: K, VK0 |
 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2024-03-27 Classification: RNA Ligands: K, VKI |
 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2024-03-27 Classification: RNA Ligands: K, NA |
 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2024-03-27 Classification: RNA Ligands: K, NA, VLR |
 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2024-03-27 Classification: RNA Ligands: K, NA, W6F |
 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2024-03-27 Classification: RNA Ligands: K, VK0 |
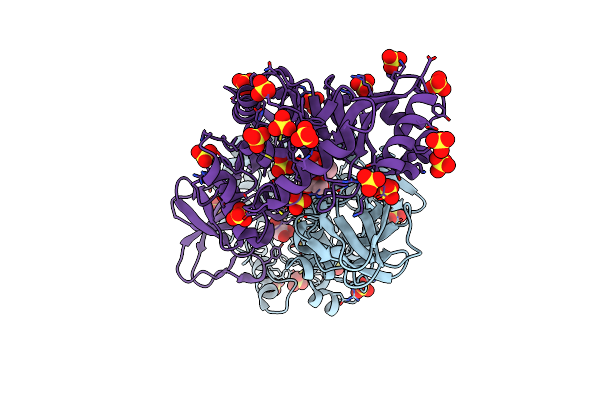 |
Organism: Staphylococcus aureus (strain mu50 / atcc 700699)
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2018-05-30 Classification: BIOSYNTHETIC PROTEIN Ligands: SO4, GOL |
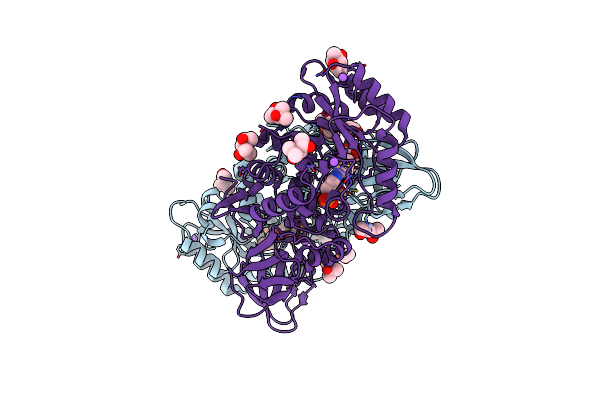 |
Structure Of The Alanine Racemase From Staphylococcus Aureus In Complex With A Pyridoxal 5' Phosphate-Derivative
Organism: Staphylococcus aureus
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2018-05-30 Classification: BIOSYNTHETIC PROTEIN Ligands: EOW, NA, MPD, ACT, CL, MRD, GOL |
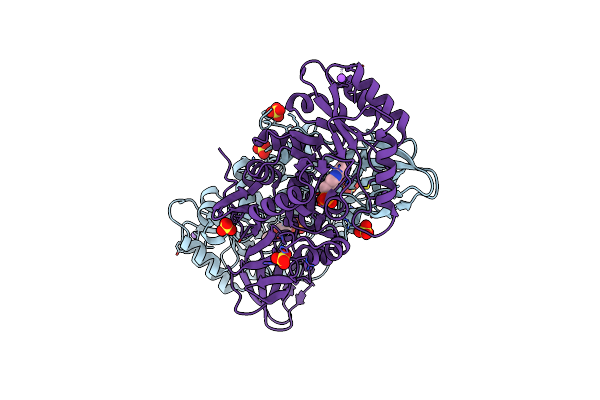 |
Structure Of The Alanine Racemase From Staphylococcus Aureus In Complex With An Pyridoxal-6- Phosphate Derivative
Organism: Staphylococcus aureus subsp. aureus mu50
Method: X-RAY DIFFRACTION Resolution:2.45 Å Release Date: 2018-05-30 Classification: BIOSYNTHETIC PROTEIN Ligands: EM2, SO4, NA, CL |
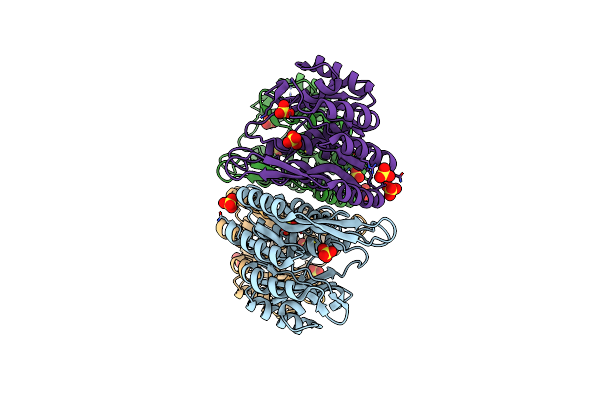 |
Organism: Staphylococcus aureus subsp. aureus mu50
Method: X-RAY DIFFRACTION Resolution:1.45 Å Release Date: 2014-03-19 Classification: TRANSFERASE Ligands: SO4 |
 |
Structure Of The Pyridoxal Kinase From Staphylococcus Aureus In Complex With Adp
Organism: Staphylococcus aureus subsp. aureus mu50
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2014-03-19 Classification: TRANSFERASE Ligands: ADP, SO4 |
 |
Structure Of The Pyridoxal Kinase From Staphylococcus Aureus In Complex With Pyridoxal
Organism: Staphylococcus aureus subsp. aureus mu50
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2014-03-19 Classification: TRANSFERASE Ligands: UEG, SO4, PXL |
 |
Structure Of The Pyridoxal Kinase From Staphylococcus Aureus In Complex With Amp-Pcp
Organism: Staphylococcus aureus subsp. aureus mu50
Method: X-RAY DIFFRACTION Resolution:1.45 Å Release Date: 2014-03-19 Classification: TRANSFERASE Ligands: ACP, SO4 |
 |
Structure Of The Pyridoxal Kinase From Staphylococcus Aureus In Complex With Amp-Pcp And Pyridoxal
Organism: Staphylococcus aureus
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2014-03-19 Classification: TRANSFERASE Ligands: PXL, ACP, SO4, UEG |

