Search Count: 134
 |
Crystal Structure Of 5'-Deoxy-5'-Methylthioadenosine Phosphorylase From Aeropyrum Pernix 353K
Organism: Aeropyrum pernix k1
Method: X-RAY DIFFRACTION Release Date: 2025-10-08 Classification: TRANSFERASE Ligands: PO4, ADE |
 |
Crystal Structure Of 5'-Deoxy-5'-Methylthioadenosine Phosphorylase From Aeropyrum Pernix Complex With 5'-Deoxy-5'-Methylthioadenosine 323K
Organism: Aeropyrum pernix k1
Method: X-RAY DIFFRACTION Release Date: 2025-10-01 Classification: TRANSFERASE Ligands: PEG, PO4, MTA |
 |
Crystal Structure Of 5'-Deoxy-5'-Methylthioadenosine Phosphorylase From Aeropyrum Pernix Complex With 5'-Deoxy-5'-Methylthioadenosine 293K
Organism: Aeropyrum pernix k1
Method: X-RAY DIFFRACTION Release Date: 2025-10-01 Classification: TRANSFERASE Ligands: PEG, PO4, MTA |
 |
Crystal Structure Of 5'-Deoxy-5'-Methylthioadenosine Phosphorylase From Aeropyrum Pernix Complex With 5'-Deoxy-5'-Methylthioadenosine 333K
Organism: Aeropyrum pernix k1
Method: X-RAY DIFFRACTION Release Date: 2025-09-17 Classification: TRANSFERASE Ligands: PO4, MTA |
 |
Crystal Structure Of 5'-Deoxy-5'-Methylthioadenosine Phosphorylase From Aeropyrum Pernix Complex With 5'-Deoxy-5'-Methylthioadenosine
Organism: Aeropyrum pernix k1
Method: X-RAY DIFFRACTION Resolution:1.22 Å Release Date: 2024-10-09 Classification: TRANSFERASE Ligands: PO4, MTA, PGE, PEG |
 |
Crystal Structure Of 5'-Deoxy-5'-Methylthioadenosine Phosphorylase From Aeropyrum Pernix Complex With 5'-Deoxy-5'-Methylthioadenosine 343K
Organism: Aeropyrum pernix k1
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2024-09-25 Classification: TRANSFERASE Ligands: PEG, PO4, MTA |
 |
Crystal Structure Of 5'-Deoxy-5'-Methylthioadenosine Phosphorylase From Aeropyrum Pernix Complex With 5'-Deoxy-5'-Methylthioadenosine 353K
Organism: Aeropyrum pernix k1
Method: X-RAY DIFFRACTION Resolution:1.62 Å Release Date: 2024-09-18 Classification: TRANSFERASE Ligands: PO4, MTA |
 |
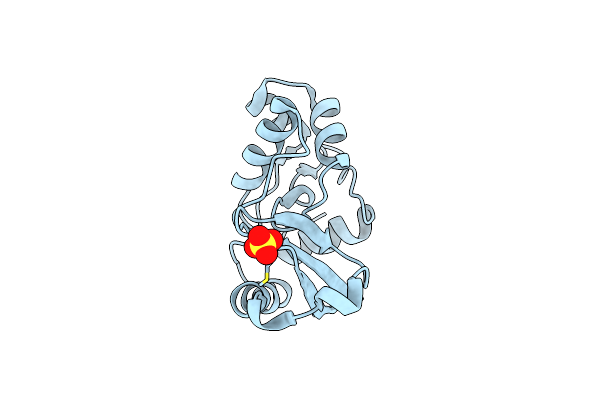
Crystal Structure Of Sulfurisphaera Tokodaii O6-Methylguanine Methyltransferase Y91F/C120S Variant In Complex With O6-Methyldeoxyguanosine
Organism: Sulfurisphaera tokodaii (strain dsm 16993 / jcm 10545 / nbrc 100140 / 7)
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2021-12-15 Classification: TRANSFERASE Ligands: J03 |
 |
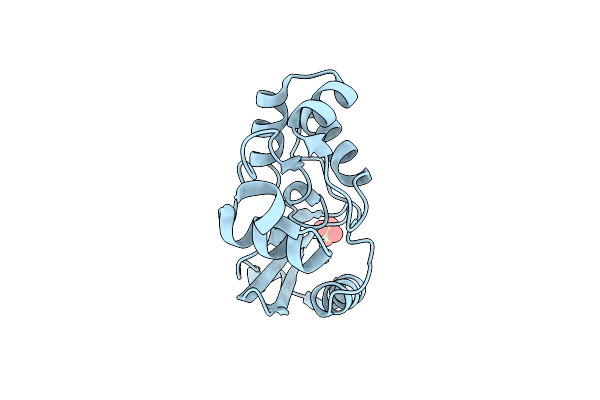
Crystal Structure Of Sulfurisphaera Tokodaii Methylated O6-Methylguanine Methyltransferase
Organism: Sulfurisphaera tokodaii (strain dsm 16993 / jcm 10545 / nbrc 100140 / 7)
Method: X-RAY DIFFRACTION Resolution:1.74 Å Release Date: 2021-12-15 Classification: TRANSFERASE Ligands: SO4 |
 |
Organism: Sulfurisphaera tokodaii (strain dsm 16993 / jcm 10545 / nbrc 100140 / 7)
Method: X-RAY DIFFRACTION Resolution:1.13 Å Release Date: 2021-12-15 Classification: TRANSFERASE Ligands: SO4 |
 |
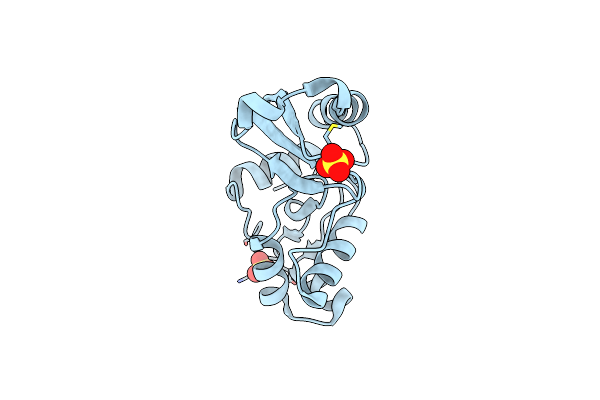
Crystal Structure Of Sulfurisphaera Tokodaii O6-Methylguanine Methyltransferase C120S Variant In Complex With O6-Methyldeoxyguanosine
Organism: Sulfurisphaera tokodaii (strain dsm 16993 / jcm 10545 / nbrc 100140 / 7)
Method: X-RAY DIFFRACTION Resolution:1.63 Å Release Date: 2021-12-15 Classification: TRANSFERASE Ligands: J03, SO4 |
 |
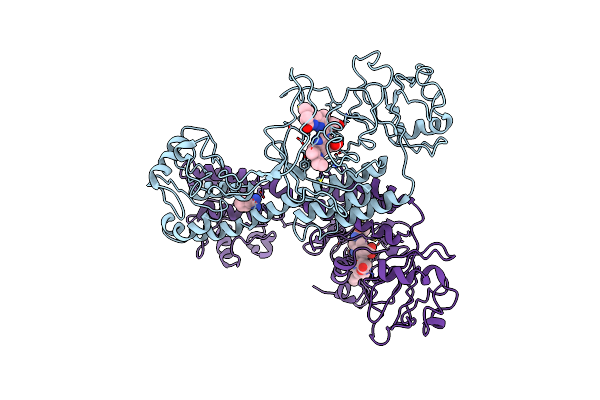
Crystal Structure Of Sulfurisphaera Tokodaii O6-Methylguanine Methyltransferase
Organism: Sulfurisphaera tokodaii (strain dsm 16993 / jcm 10545 / nbrc 100140 / 7)
Method: X-RAY DIFFRACTION Resolution:1.79 Å Release Date: 2021-12-01 Classification: TRANSFERASE Ligands: SO4 |
 |
Organism: Stigmatella aurantiaca (strain dw4/3-1)
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2021-10-06 Classification: SIGNALING PROTEIN Ligands: VHG, BEN |
 |
Crystal Structure Of Sulfurisphaera Tokodaii O6-Methylguanine Methyltransferase Y91F/C120S Variant
Organism: Sulfurisphaera tokodaii (strain dsm 16993 / jcm 10545 / nbrc 100140 / 7)
Method: X-RAY DIFFRACTION Resolution:1.78 Å Release Date: 2021-09-29 Classification: TRANSFERASE Ligands: SO4 |
 |
Crystal Structure Of Sulfurisphaera Tokodaii O6-Methylguanine Methyltransferase C120S Variant
Organism: Sulfurisphaera tokodaii (strain dsm 16993 / jcm 10545 / nbrc 100140 / 7)
Method: X-RAY DIFFRACTION Resolution:1.25 Å Release Date: 2021-08-18 Classification: TRANSFERASE Ligands: SO4 |
 |
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2021-05-12 Classification: VIRAL PROTEIN, Hydrolase Ligands: ASC, ETF |
 |
High-Resolution Crystal Structures Of Transient Intermediates In The Phytochrome Photocycle, 33 Ms Structure
Organism: Stigmatella aurantiaca
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2021-04-07 Classification: SIGNALING PROTEIN Ligands: 3Q8, BEN |
 |
Crystal Structure Of Cystathionine Gamma-Lyase From Lactobacillus Plantarum Complexed With L-Serine
Organism: Lactobacillus plantarum
Method: X-RAY DIFFRACTION Resolution:2.75 Å Release Date: 2020-10-07 Classification: LYASE Ligands: KOU, PO4 |
 |
Crystal Structure Of Cystathionine Gamma-Lyase From Lactobacillus Plantarum Complexed With Cystathionine
Organism: Lactobacillus plantarum
Method: X-RAY DIFFRACTION Resolution:3.10 Å Release Date: 2020-10-07 Classification: LYASE Ligands: E9U, PO4 |
 |
Organism: Stigmatella aurantiaca dw4/3-1
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2019-10-09 Classification: SIGNALING PROTEIN Ligands: EL5, BEN |

