Search Count: 26
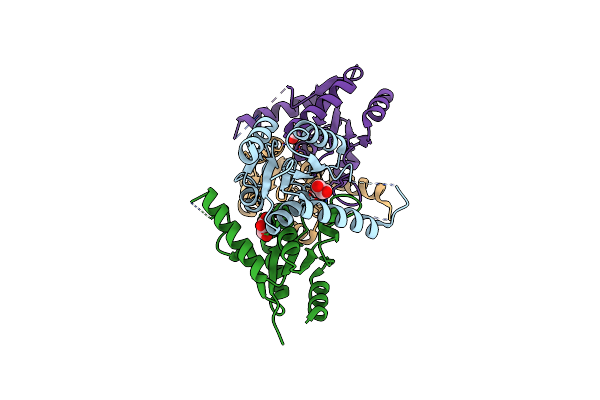 |
Crystal Structure Of A 2`-Deoxyribosyltransferase From The Psychrophilic Bacterium Desulfotalea Psychrophila.
Organism: Desulfotalea psychrophila (strain lsv54 / dsm 12343)
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2021-10-20 Classification: TRANSFERASE Ligands: GOL |
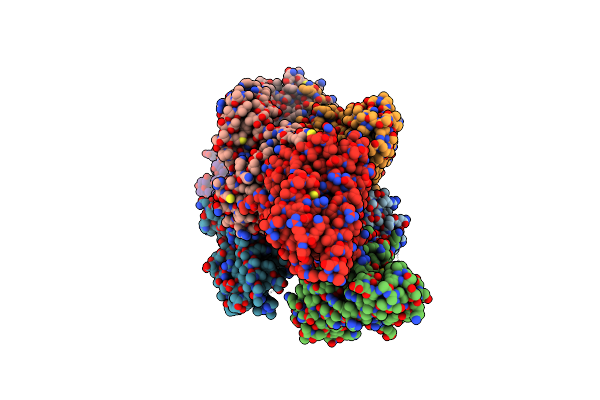 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2021-07-21 Classification: SUGAR BINDING PROTEIN Ligands: M6P, ACT |
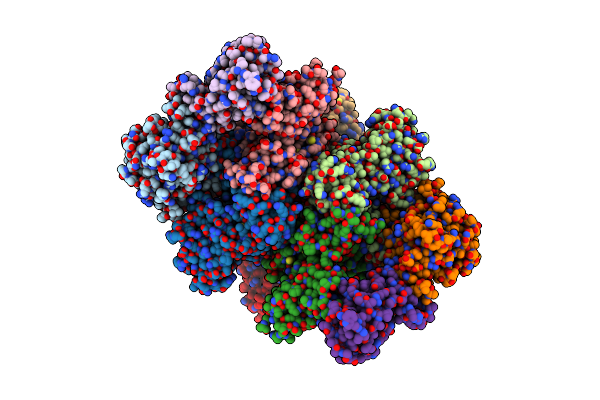 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2021-07-14 Classification: TRANSLATION Ligands: F6P |
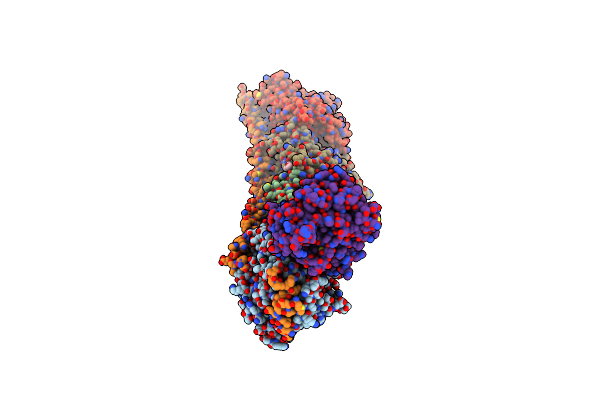 |
Organism: Rattus norvegicus, Gallus gallus, Sus scrofa
Method: X-RAY DIFFRACTION Resolution:3.34 Å Release Date: 2021-03-31 Classification: LIGASE Ligands: GTP, MG, CA, GDP, MES, O9B, O9H, OH5, PGE, VAL, ACP |
 |
Organism: Rattus norvegicus, Gallus gallus, Sus scrofa
Method: X-RAY DIFFRACTION Resolution:2.85 Å Release Date: 2021-03-31 Classification: LIGASE Ligands: GTP, MG, CA, GDP, MES, O9B, O9K, O9N, PGE, VAL, P6S, PEG, ACP |
 |
Organism: Deinococcus radiodurans (strain atcc 13939 / dsm 20539 / jcm 16871 / lmg 4051 / nbrc 15346 / ncimb 9279 / r1 / vkm b-1422)
Method: X-RAY DIFFRACTION Resolution:1.81 Å Release Date: 2019-01-16 Classification: TRANSFERASE Ligands: ATP, MG, GOL, MPD, CL |
 |
Organism: Xanthobacter autotrophicus py2
Method: X-RAY DIFFRACTION Resolution:1.87 Å Release Date: 2017-08-09 Classification: LIGASE Ligands: MN, ACT, AE4, MG, AMP, ZN |
 |
Mechanism Of Atp-Dependent Acetone Carboxylation, Acetone Carboxylase Amp Bound Structure
Organism: Xanthobacter autotrophicus (strain atcc baa-1158 / py2)
Method: X-RAY DIFFRACTION Resolution:2.65 Å Release Date: 2017-08-09 Classification: LIGASE Ligands: MN, AMP, SO4, ZN |
 |
Mechanism Of Atp-Dependent Acetone Carboxylation, Acetone Carboxylase Nucleotide-Free Structure
Organism: Xanthobacter autotrophicus (strain atcc baa-1158 / py2)
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2017-08-09 Classification: LIGASE Ligands: MN, SO4, ZN |
 |
Crystal Structure Of A Five-Domain Gh115 Alpha-Glucuronidase From The Marine Bacterium Saccharophagus Degradans 2-40T
Organism: Saccharophagus degradans
Method: X-RAY DIFFRACTION Resolution:1.93 Å Release Date: 2016-05-04 Classification: HYDROLASE Ligands: PO4, NA, GOL, ACT |
 |
Central Domain Of Uncharacterized Lpg1148 Protein From Legionella Pneumophila
Organism: Legionella pneumophila subsp. pneumophila
Method: X-RAY DIFFRACTION Resolution:1.93 Å Release Date: 2015-09-16 Classification: UNKNOWN FUNCTION Ligands: CL |
 |
Organism: Chryseobacterium sp. strb126
Method: X-RAY DIFFRACTION Resolution:1.09 Å Release Date: 2015-07-15 Classification: HYDROLASE Ligands: ZN |
 |
Aidc, A Dizinc Quorum-Quenching Lactonase, In Complex With A Product N-Hexnoyl-L-Homoserine
Organism: Chryseobacterium sp. strb126
Method: X-RAY DIFFRACTION Resolution:1.67 Å Release Date: 2015-07-15 Classification: HYDROLASE Ligands: ZN, C6L |
 |
Structure Of C-Terminal Domain Of Uncharacterized Protein From Legionella Pneumophila
Organism: Legionella pneumophila subsp. pneumophila str. philadelphia 1
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2015-05-06 Classification: STRUCTURAL GENOMICS, UNKNOWN FUNCTION |
 |
The Crystal Structure Of The N-Terminal Fragment Of Uncharacterized Protein From Legionella Pneumophila
Organism: Legionella pneumophila subsp. pneumophila str. philadelphia 1
Method: X-RAY DIFFRACTION Resolution:1.10 Å Release Date: 2015-04-08 Classification: STRUCTURAL GENOMICS, UNKNOWN FUNCTION |
 |
Crystal Structure Of The Complex Between The N-Terminal Domain Of Ravj And Legl1 From Legionella Pneumophila Str. Philadelphia
Organism: Legionella pneumophila subsp. pneumophila
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2015-01-28 Classification: Structural genomics, Unknown Function |
 |
Crystal Structure Of Gh62 Hydrolase From Thermophilic Fungus Scytalidium Thermophilum
Organism: Scytalidium thermophilum
Method: X-RAY DIFFRACTION Resolution:1.23 Å Release Date: 2014-11-19 Classification: HYDROLASE Ligands: GOL, PO4 |
 |
Crystal Structure Of Lpg1851 Protein From Legionella Pneumophila (Putative T4Ss Effector)
Organism: Legionella pneumophila subsp. pneumophila
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2012-11-07 Classification: Structural Genomics, Unknown Function Ligands: CIT, SIN |
 |
Crystal Structure Of Mono-Zinc Form Of Succinyl-Diaminopimelate Desuccinylase From Haemophilus Influenzae
Organism: Haemophilus influenzae
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2009-11-10 Classification: HYDROLASE Ligands: ZN, SO4 |
 |
Crystal Structure Of Zinc-Bound Succinyl-Diaminopimelate Desuccinylase From Haemophilus Influenzae
Organism: Haemophilus influenzae
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2009-09-22 Classification: HYDROLASE Ligands: ZN, GOL, SO4 |

