Search Count: 35
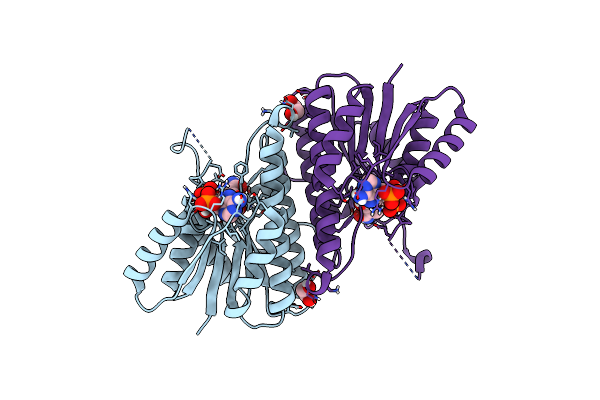 |
Organism: Clostridium acetobutylicum
Method: X-RAY DIFFRACTION Release Date: 2025-07-02 Classification: OXIDOREDUCTASE Ligands: NAP, EDO |
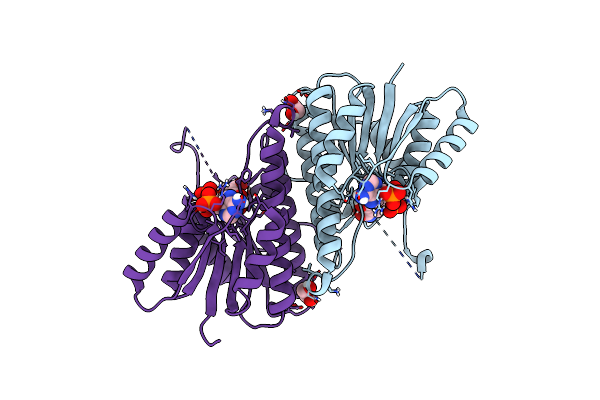 |
Clostridium Acetobutylicum Alcohol Dehydrogenase Bound To Nadp+, Disordered Nicotinamide
Organism: Clostridium acetobutylicum
Method: X-RAY DIFFRACTION Release Date: 2025-07-02 Classification: OXIDOREDUCTASE Ligands: NAP, EDO |
 |
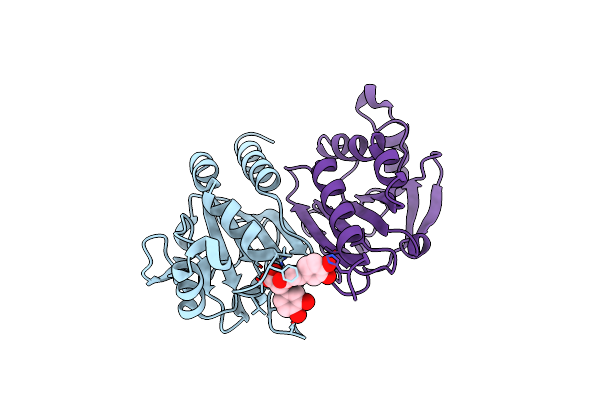
Organism: Alcaligenes faecalis
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2025-02-26 Classification: METAL BINDING PROTEIN Ligands: ZN |
 |
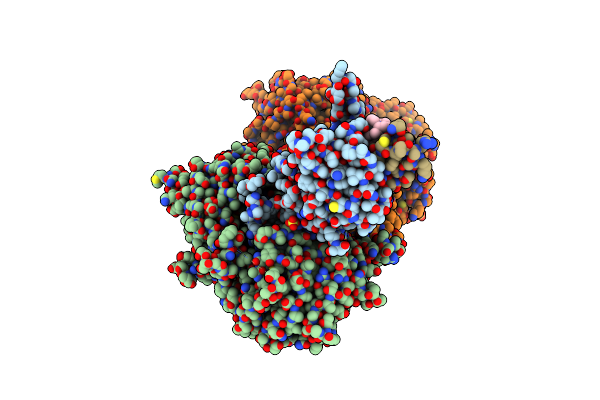
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.71 Å Release Date: 2025-01-22 Classification: OXIDOREDUCTASE Ligands: ROA |
 |
Ternary Complex Structure Of Cereblon-Ddb1 Bound To Wiz(Zf7) And The Molecular Glue Dwiz-1
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.15 Å Release Date: 2024-07-10 Classification: LIGASE Ligands: ZN, U3I, SO4, EDO |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2022-12-14 Classification: OXIDOREDUCTASE Ligands: FAD, AU |
 |
Organism: Enterovirus a71
Method: X-RAY DIFFRACTION Release Date: 2022-06-22 Classification: VIRAL PROTEIN Ligands: G7L |
 |
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Release Date: 2022-06-22 Classification: VIRAL PROTEIN Ligands: G7L |
 |
Organism: Enterovirus a71
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2022-06-22 Classification: VIRAL PROTEIN Ligands: G7F |
 |
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2022-06-22 Classification: VIRAL PROTEIN Ligands: G7O |
 |
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:2.75 Å Release Date: 2021-11-03 Classification: VIRAL PROTEIN Ligands: AU |
 |
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:1.72 Å Release Date: 2021-11-03 Classification: VIRAL PROTEIN Ligands: AU |
 |
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:1.77 Å Release Date: 2021-11-03 Classification: VIRAL PROTEIN |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.78 Å Release Date: 2020-09-02 Classification: LIGASE Ligands: TYS, PG4, NA, SO4 |
 |
Organism: Roseiflexus castenholzii
Method: ELECTRON MICROSCOPY Release Date: 2018-05-02 Classification: PHOTOSYNTHESIS Ligands: BCL, BPH, MQE, HEM, KGD, FE |
 |
Crystal Structure Of Human T2-Tryptophanyl-Trna Synthetase With H130R Mutation
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.79 Å Release Date: 2018-01-17 Classification: LIGASE |
 |
Crystal Structure Of Human H130R Tryptophanyl-Trna Synthetase In Complex With Trpamp
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2018-01-17 Classification: LIGASE Ligands: TYM, MG |
 |
Structure Of Prethrombin-2 Mutant S195A Bound To The Active Site Inhibitor Argatroban
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2014-11-05 Classification: HYDROLASE/HYDROLASE INHIBITOR Ligands: 15U |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.30 Å Release Date: 2013-06-26 Classification: HYDROLASE Ligands: NAG |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.19 Å Release Date: 2013-03-13 Classification: HYDROLASE/PEPTIDE |

