Search Count: 15
 |
Organism: Arabidopsis thaliana
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2020-06-10 Classification: OXIDOREDUCTASE Ligands: HEM, GOL, SOG |
 |
Organism: Arabidopsis thaliana
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2020-06-10 Classification: OXIDOREDUCTASE Ligands: HEM |
 |
Crystal Structure Of Cyp97A3 Mutant S290D/W300L/S304V In Complex With Retinal
Organism: Arabidopsis thaliana
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2020-06-10 Classification: OXIDOREDUCTASE Ligands: HEM, RET |
 |
Organism: Arabidopsis thaliana
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2020-01-22 Classification: OXIDOREDUCTASE Ligands: HEM |
 |
Organism: Arabidopsis thaliana
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2020-01-22 Classification: OXIDOREDUCTASE Ligands: HEM, RET |
 |
Organism: Arabidopsis thaliana
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2017-01-25 Classification: OXIDOREDUCTASE Ligands: FMN, FAD |
 |
Organism: Synechocystis sp. pcc 6803 substr. kazusa, Escherichia coli bl21(de3)
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2016-09-21 Classification: HYDROLASE Ligands: ZN, NA |
 |
Organism: Synechocystis sp. pcc 6803 substr. kazusa, Escherichia coli bl21(de3)
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2016-09-21 Classification: HYDROLASE Ligands: ZN, NA |
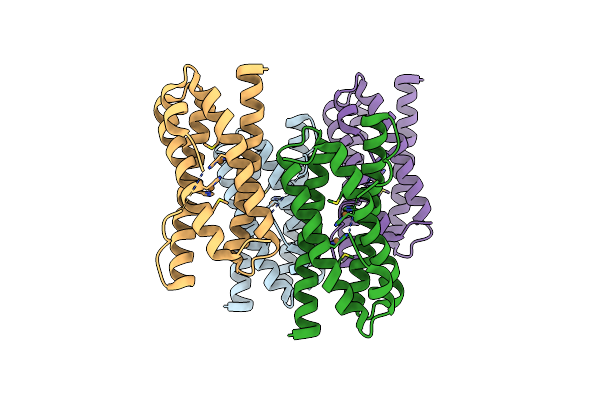 |
Organism: Synechocystis sp. pcc 6803
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2016-09-07 Classification: METAL BINDING PROTEIN Ligands: CU1 |
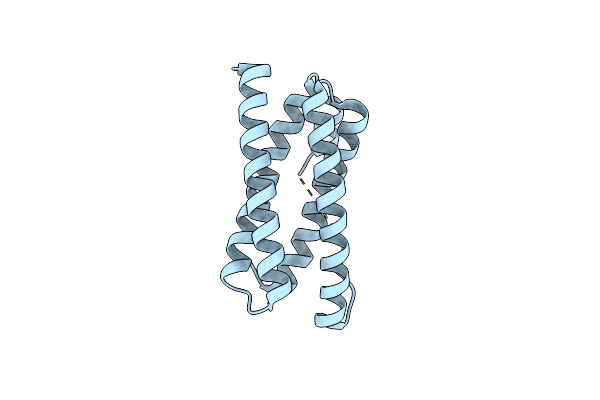 |
Organism: Synechocystis sp. pcc 6803
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2016-09-07 Classification: METAL BINDING PROTEIN |
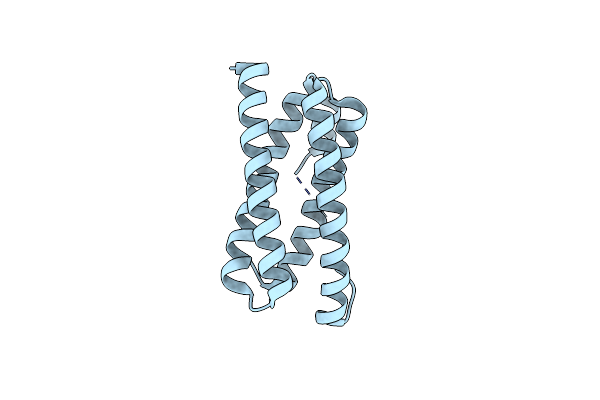 |
Organism: Synechocystis sp. pcc 6803
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2016-09-07 Classification: METAL BINDING PROTEIN |
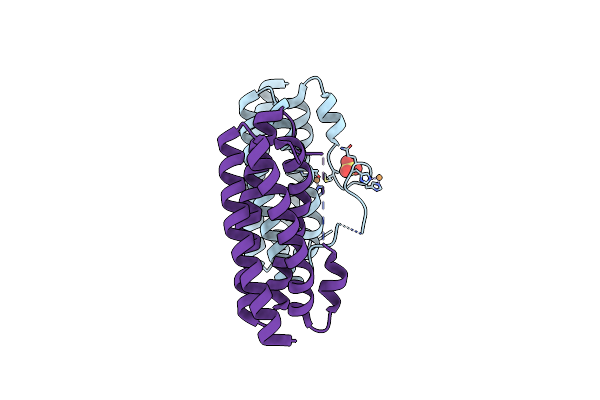 |
Organism: Synechocystis sp. pcc 6803
Method: X-RAY DIFFRACTION Resolution:2.01 Å Release Date: 2016-09-07 Classification: METAL BINDING PROTEIN Ligands: CU, SO4 |
 |
Organism: Synechocystis sp. pcc 6803
Method: X-RAY DIFFRACTION Resolution:1.45 Å Release Date: 2016-09-07 Classification: METAL BINDING PROTEIN Ligands: AG |
 |
Organism: Synechocystis sp. pcc 6803
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2016-09-07 Classification: METAL BINDING PROTEIN Ligands: AG |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2011-06-01 Classification: HYDROLASE Ligands: ZN |

