Search Count: 22
 |
Characterization Of Solanum Tuberosum Multicystatin And Significance Of Core Domains
Organism: Solanum tuberosum
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2014-02-26 Classification: HYDROLASE INHIBITOR |
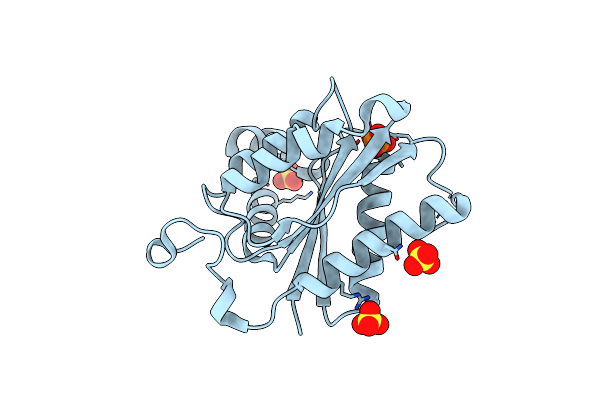 |
Organism: Edta-degrading bacterium bnc1
Method: X-RAY DIFFRACTION Resolution:2.19 Å Release Date: 2013-08-07 Classification: OXIDOREDUCTASE Ligands: PO4, SO4 |
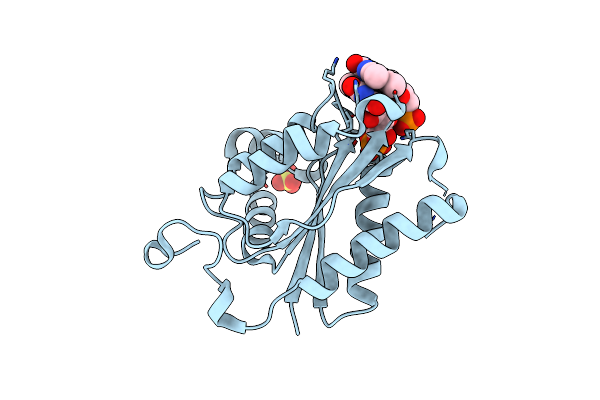 |
Organism: Edta-degrading bacterium bnc1
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2013-08-07 Classification: OXIDOREDUCTASE Ligands: FMN, SO4 |
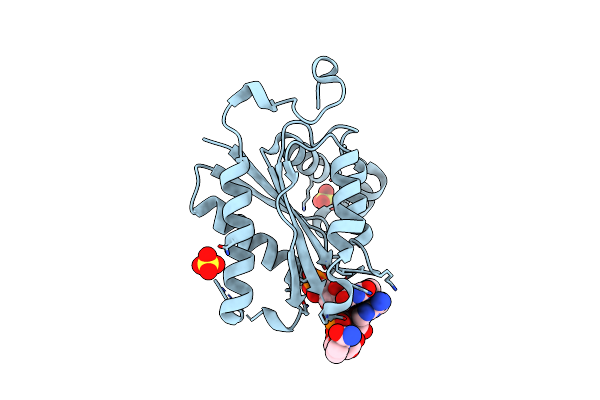 |
Organism: Edta-degrading bacterium bnc1
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2013-08-07 Classification: OXIDOREDUCTASE Ligands: NAI, FMN, SO4 |
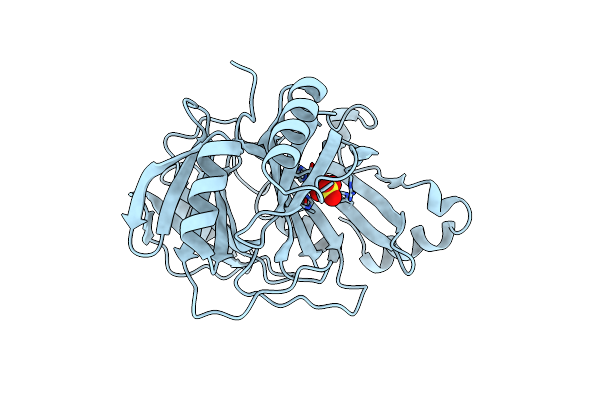 |
Organism: Sphingobium chlorophenolicum
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2013-04-17 Classification: OXIDOREDUCTASE Ligands: FE, SO4 |
 |
Organism: Ralstonia eutropha
Method: X-RAY DIFFRACTION Resolution:1.76 Å Release Date: 2012-06-13 Classification: OXIDOREDUCTASE Ligands: ZN, GOL, SO4, NAD |
 |
Organism: Ralstonia eutropha
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2012-06-13 Classification: OXIDOREDUCTASE Ligands: ZN, ISP, SO4, NAD, FU2 |
 |
Organism: Ralstonia eutropha
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2012-06-13 Classification: OXIDOREDUCTASE Ligands: ZN, FU2, SO4, NAD |
 |
Organism: Ralstonia eutropha
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2012-06-13 Classification: OXIDOREDUCTASE Ligands: ZN, FU2, SO4, NAD |
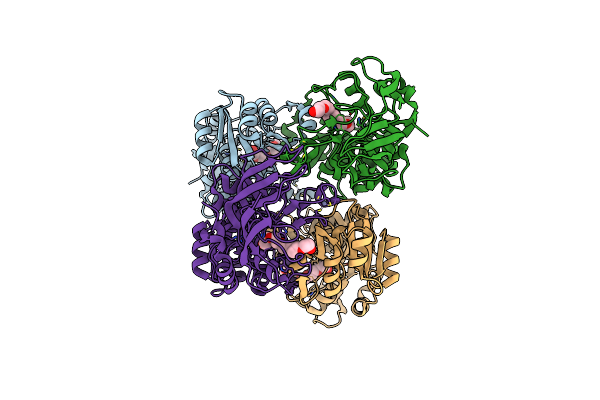 |
Organism: Ralstonia eutropha
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2012-04-25 Classification: OXIDOREDUCTASE Ligands: ZN, P6G |
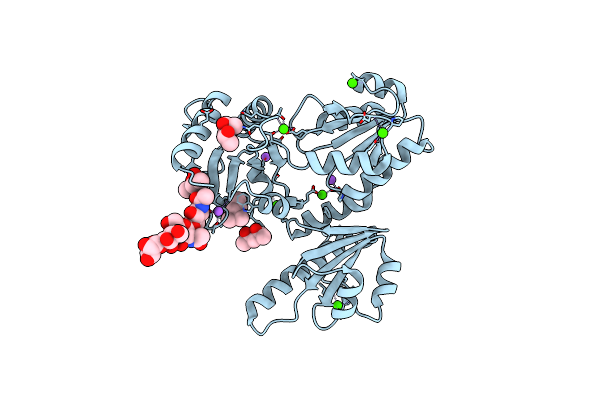 |
Organism: Oryctolagus cuniculus
Method: X-RAY DIFFRACTION Resolution:1.76 Å Release Date: 2011-12-21 Classification: Calcium-binding protein Ligands: MRD, MPD, CA, NA |
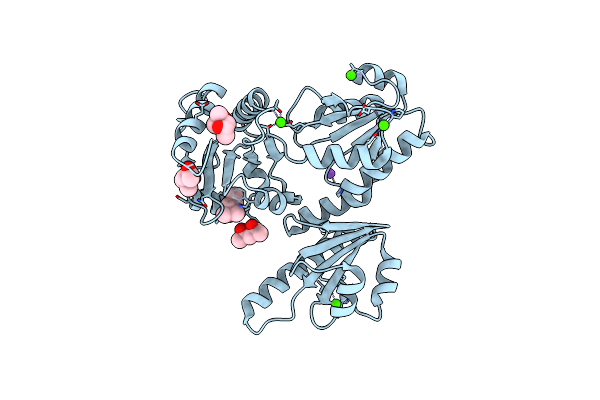 |
Organism: Oryctolagus cuniculus
Method: X-RAY DIFFRACTION Resolution:1.74 Å Release Date: 2011-12-21 Classification: calcium-binding protein Ligands: MPD, MRD, CA, NA |
 |
Organism: Solanum tuberosum
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2010-02-02 Classification: HYDROLASE INHIBITOR |
 |
Organism: Solanum tuberosum
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2010-02-02 Classification: HYDROLASE INHIBITOR |
 |
Crystal Structure Of Chlorophenol 4-Monooxygenase (Tftd) Of Burkholderia Cepacia Ac1100
Organism: Burkholderia cepacia
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2009-11-10 Classification: OXIDOREDUCTASE |
 |
Organism: Burkholderia cepacia
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2009-11-10 Classification: OXIDOREDUCTASE |
 |
Organism: Burkholderia cepacia
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2009-11-10 Classification: OXIDOREDUCTASE Ligands: FAD |
 |
Organism: Burkholderia cepacia
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2009-11-10 Classification: OXIDOREDUCTASE Ligands: FAD, NAD |
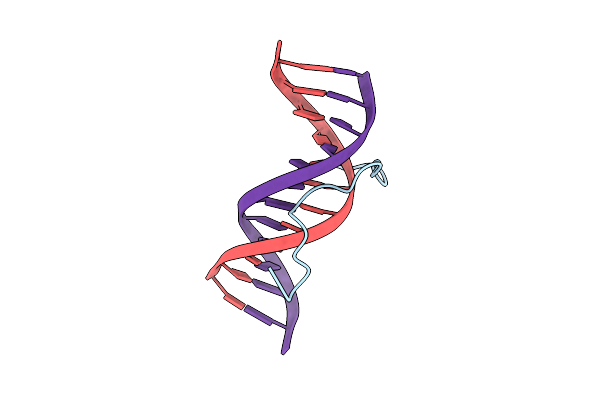 |
Solution Structure Of A Complex Of The Second Dna Binding Domain Of Human Hmg-I(Y) Bound To Dna Dodecamer Containing The Prdii Site Of The Interferon-Beta Promoter, Nmr, Minimized Average Structure
Organism: Homo sapiens
Method: SOLUTION NMR Release Date: 1997-10-15 Classification: DNA BINDING PROTEIN/DNA |
 |
Solution Structure Of A Complex Of The Second Dna Binding Domain Of Human Hmg-I(Y) Bound To Dna Dodecamer Containing The Prdii Site Of The Interferon-Beta Promoter, Nmr, 35 Structures
Organism: Homo sapiens
Method: SOLUTION NMR Release Date: 1997-10-15 Classification: DNA BINDING PROTEIN/DNA |

