Search Count: 82
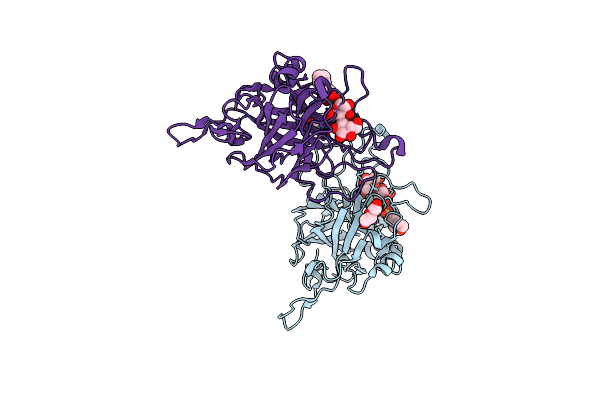 |
Coprinopsis Cinerea Gh131 Protein Ccgh131B E161A In Complex With Cellobiose
Organism: Coprinopsis cinerea (strain okayama-7 / 130 / atcc mya-4618 / fgsc 9003)
Method: X-RAY DIFFRACTION Release Date: 2024-11-06 Classification: HYDROLASE Ligands: MES, PEG, PGE |
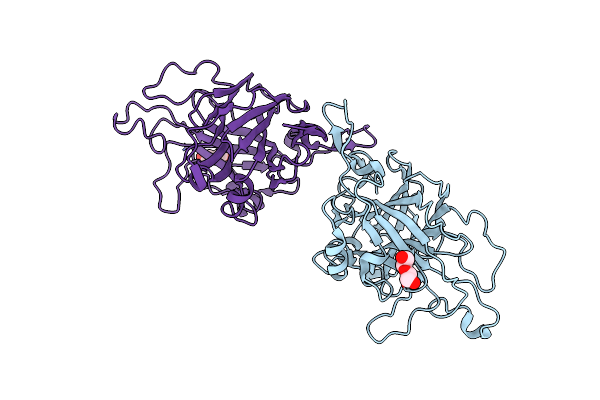 |
Organism: Coprinopsis cinerea (strain okayama-7 / 130 / atcc mya-4618 / fgsc 9003)
Method: X-RAY DIFFRACTION Release Date: 2024-11-06 Classification: HYDROLASE Ligands: PEG |
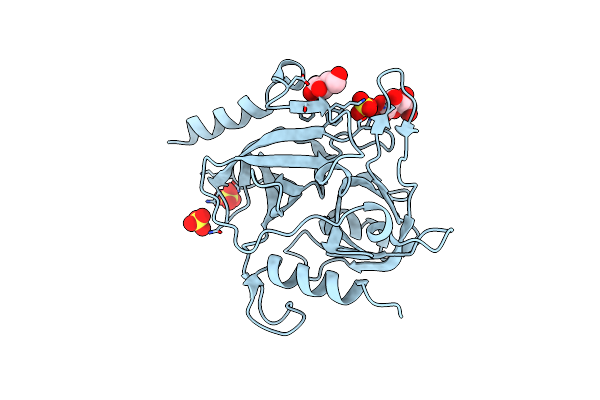 |
Organism: Staphylococcus aureus
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2024-09-11 Classification: HYDROLASE Ligands: SO4, BGC |
 |
Organism: Rhodothermus marinus jcm 9785
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2024-02-07 Classification: HYDROLASE Ligands: MN, CA, MES, MPD |
 |
Rhodothermus Marinus Alpha-Amylase Rmgh13_47A Cbm48-A-B-C Domains In Complex With Branched Pentasaccharide
Organism: Rhodothermus marinus jcm 9785
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2024-02-07 Classification: HYDROLASE Ligands: MN, CA, MES, PGE, PEG, GOL |
 |
Organism: Beijerinckia indica subsp. indica
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2023-06-14 Classification: TRANSFERASE Ligands: GOL |
 |
Beijerinckia Indica Beta-Fructosyltransferase Variant H395R/F473Y In Complex With Fructose
Organism: Beijerinckia indica subsp. indica nbrc 3744
Method: X-RAY DIFFRACTION Resolution:1.36 Å Release Date: 2023-06-14 Classification: TRANSFERASE Ligands: MG, FRU, BDF |
 |
Organism: Aspergillus sojae nbrc 4239
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2022-06-15 Classification: HYDROLASE Ligands: NAG, NA, GLC |
 |
Organism: Frischella perrara
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2022-03-09 Classification: HYDROLASE Ligands: PEG, PGE |
 |
Organism: Frischella perrara
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2022-03-09 Classification: HYDROLASE Ligands: FRU, PEG, 1PE |
 |
Organism: Beijerinckia indica subsp. indica (strain atcc 9039 / dsm 1715 / ncib 8712)
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2020-08-12 Classification: HYDROLASE Ligands: MG |
 |
Organism: Beijerinckia indica subsp. indica (strain atcc 9039 / dsm 1715 / ncib 8712)
Method: X-RAY DIFFRACTION Resolution:1.35 Å Release Date: 2020-08-12 Classification: HYDROLASE Ligands: MG, FRU, BDF |
 |
Organism: Flavobacterium johnsoniae (strain atcc 17061 / dsm 2064 / uw101)
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2019-04-24 Classification: HYDROLASE Ligands: ACT, MPD |
 |
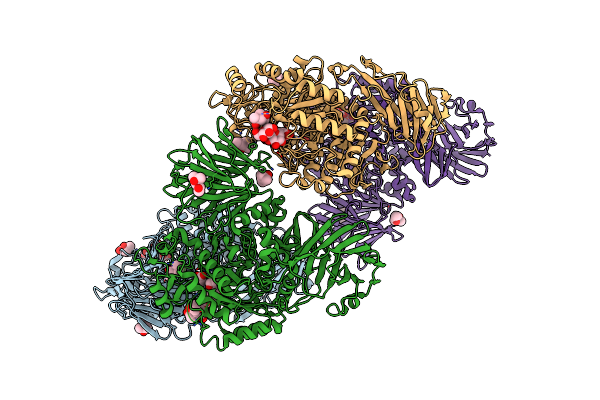
Flavobacterium Johnsoniae Gh31 Dextranase, Fjdex31A, Complexed With Glucose
Organism: Flavobacterium johnsoniae (strain atcc 17061 / dsm 2064 / uw101)
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2019-04-24 Classification: HYDROLASE Ligands: ACT, MPD |
 |
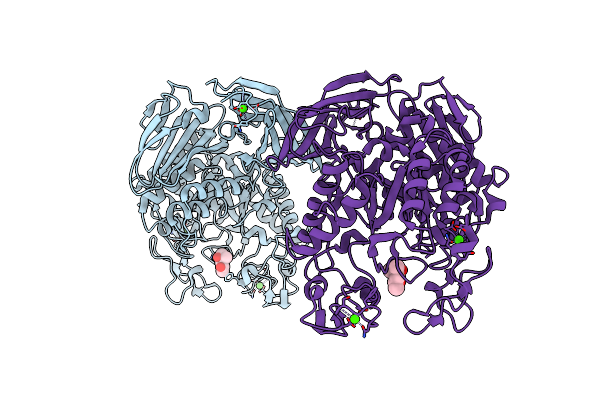
Flavobacterium Johnsoniae Gh31 Dextranase, Fjdex31A, Mutant D412A Complexed With Isomaltotriose
Organism: Flavobacterium johnsoniae (strain atcc 17061 / dsm 2064 / uw101)
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2019-04-24 Classification: HYDROLASE Ligands: ACT, MPD |
 |
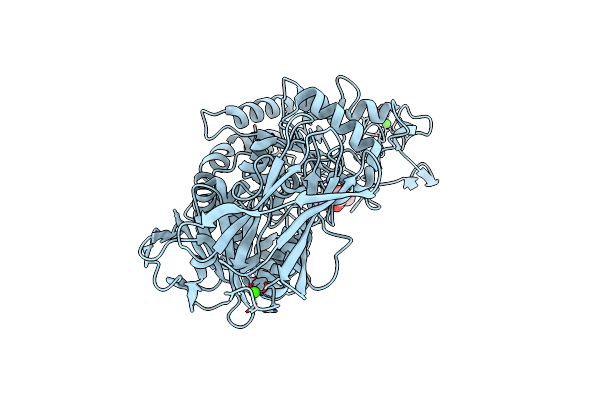
Thermoactinomyces Vulgaris R-47 Alpha-Amylase I (Tva I) Mutant A357V/Q359N/Y360E (Aqy/Vne)
Organism: Thermoactinomyces vulgaris
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2018-02-21 Classification: HYDROLASE Ligands: CA, MPD |
 |
Thermoactinomyces Vulgaris R-47 Alpha-Amylase I (Tva I) 11 Residues (From A363 To N373) Deletion Mutant (Del11)
Organism: Thermoactinomyces vulgaris
Method: X-RAY DIFFRACTION Resolution:1.37 Å Release Date: 2018-02-21 Classification: HYDROLASE Ligands: CA, MPD |
 |
Organism: Aspergillus kawachii (strain nbrc 4308)
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2017-07-19 Classification: HYDROLASE |
 |
Organism: Aspergillus kawachii (strain nbrc 4308)
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2017-07-19 Classification: HYDROLASE |
 |
Organism: Aspergillus kawachii (strain nbrc 4308)
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2017-07-19 Classification: HYDROLASE |

