Search Count: 14
 |
Organism: Thermus thermophilus hb8
Method: ELECTRON MICROSCOPY Resolution:2.80 Å Release Date: 2024-12-04 Classification: MOTOR PROTEIN |
 |
Organism: Thermus thermophilus hb8
Method: ELECTRON MICROSCOPY Release Date: 2024-12-04 Classification: MOTOR PROTEIN |
 |
Organism: Thermus thermophilus hb8
Method: ELECTRON MICROSCOPY Release Date: 2024-12-04 Classification: MOTOR PROTEIN |
 |
Organism: Thermus thermophilus hb8
Method: ELECTRON MICROSCOPY Release Date: 2024-12-04 Classification: MOTOR PROTEIN |
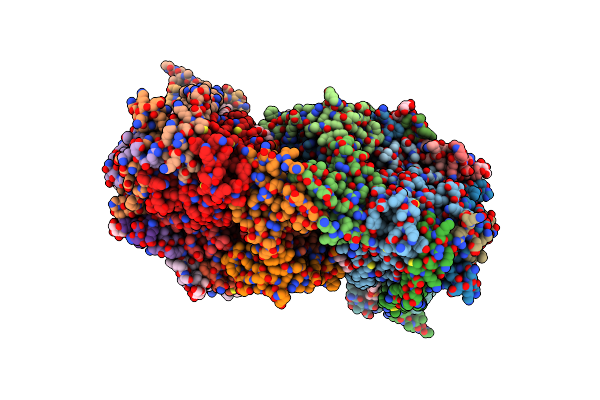 |
Crystal Structure Of Bovine Heart Cytochrome C Oxidase, Apo Structure With Dmso
Organism: Bos taurus
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2022-12-21 Classification: OXIDOREDUCTASE Ligands: HEA, CU, MG, NA, PER, PGV, TGL, CUA, CDL, CHD, PEK, PSC, ZN, DMU |
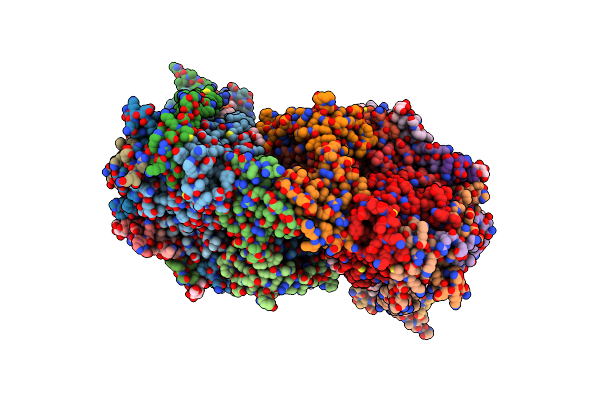 |
Crystal Structure Of Bovine Heart Cytochrome C Oxidase, The Structure Complexed With An Allosteric Inhibitor T113
Organism: Bos taurus
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2022-12-21 Classification: OXIDOREDUCTASE Ligands: HEA, CU, MG, NA, PER, TGL, J6X, CUA, CHD, PSC, PEK, PGV, CDL, DMU, ZN |
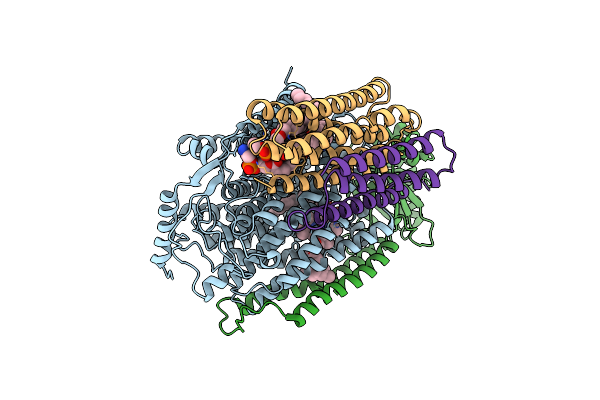 |
Cryo-Em Structure Of Cytochrome Bo3 From Escherichia Coli, Apo Structure With Dmso
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2022-12-21 Classification: OXIDOREDUCTASE Ligands: HEO, HEM, CU, PEE, UNX |
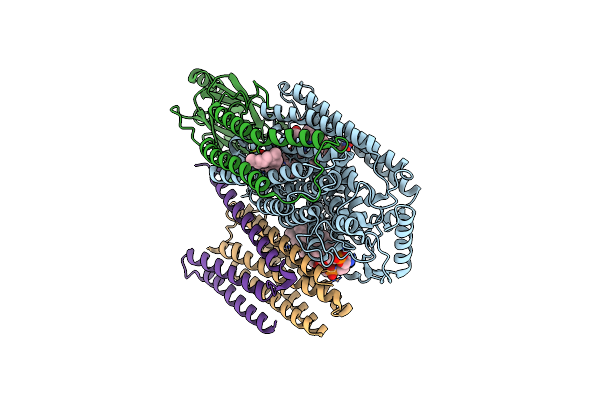 |
Cryo-Em Structure Of Cytochrome Bo3 From Escherichia Coli, The Structure Complexed With An Allosteric Inhibitor N4
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2022-12-21 Classification: OXIDOREDUCTASE Ligands: HEO, HEM, CU, PEE, JYR, UNX |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2019-05-08 Classification: HYDROLASE Ligands: ONC |
 |
The Crystal Structure Of An Archaeal Cpsf Subunit, Ph1404 From Pyrococcus Horikoshii
Organism: Pyrococcus horikoshii
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2010-04-21 Classification: HYDROLASE Ligands: SO4, ACY, ZN |
 |
The Crystal Structure Of An Archaeal Cpsf Subunit, Ph1404 From Pyrococcus Horikoshii Complexed With Rna-Analog
Organism: Pyrococcus horikoshii
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2010-04-21 Classification: HYDROLASE/RNA Ligands: SO4, ZN |
 |
Organism: Aquifex aeolicus
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2009-07-21 Classification: HYDROLASE Ligands: CL, EDO, PEG, TRS, PGE, PG4 |
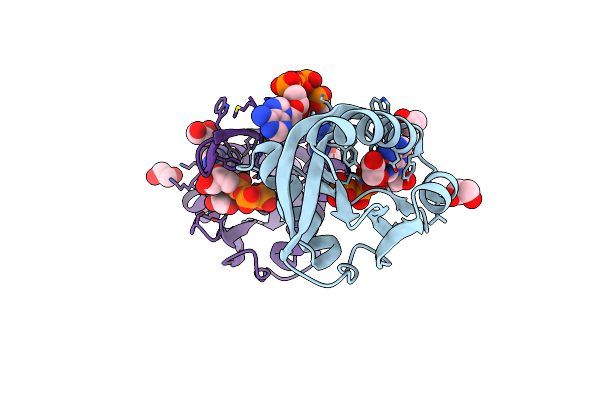 |
Crystal Structure Of Ap4A Hydrolase Complexed With Ap4A (Atp) (Aq_158) From Aquifex Aeolicus Vf5
Organism: Aquifex aeolicus vf5
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2009-07-21 Classification: HYDROLASE Ligands: ATP, B4P, EDO, GOL |
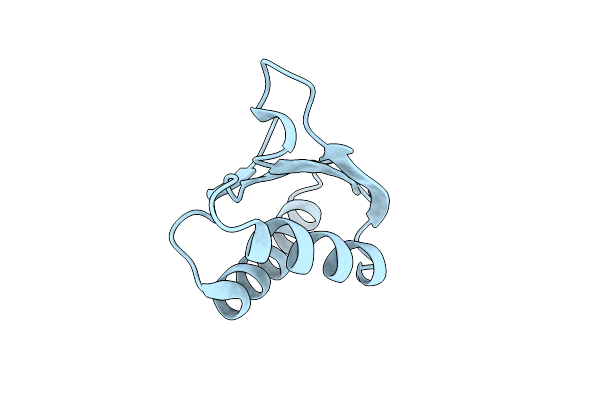 |
Organism: Thermus thermophilus
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2008-09-30 Classification: DNA BINDING PROTEIN |

