Search Count: 100
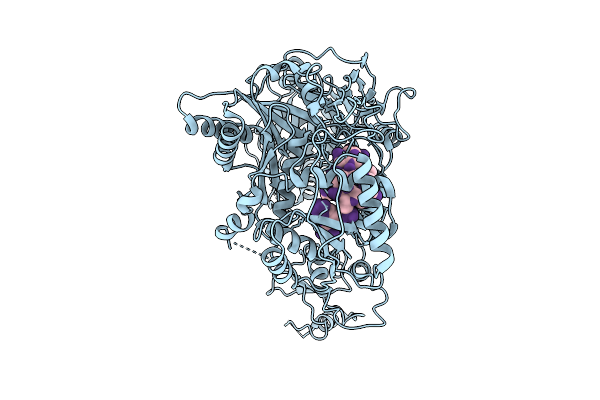 |
Structure Of Neurodevelopmental Mutant Ago1 F180Del In Complex With Guide Rna
Organism: Homo sapiens, Trichoplusia ni
Method: ELECTRON MICROSCOPY Release Date: 2025-11-05 Classification: GENE REGULATION |
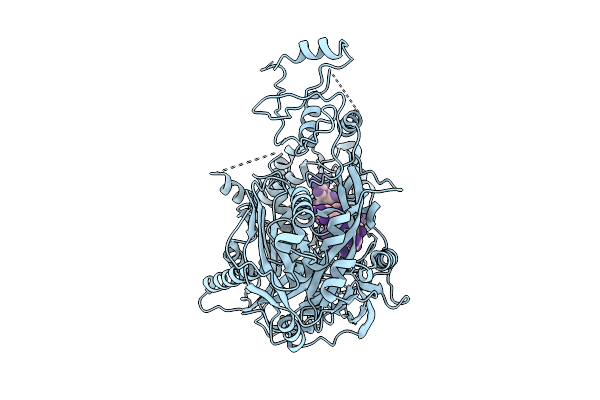 |
Organism: Homo sapiens, Trichoplusia ni
Method: ELECTRON MICROSCOPY Release Date: 2025-10-29 Classification: GENE REGULATION |
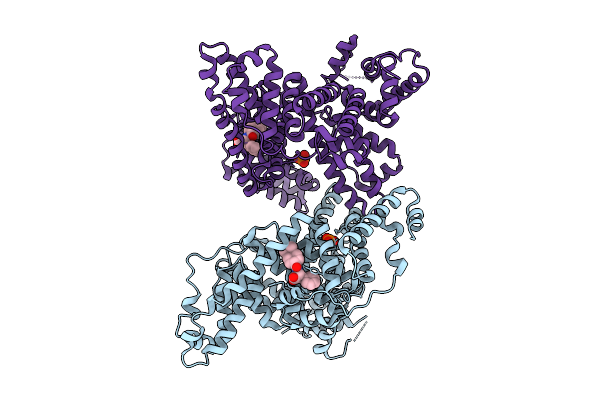 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-07-16 Classification: TRANSPORT PROTEIN Ligands: A1L9H, PO4 |
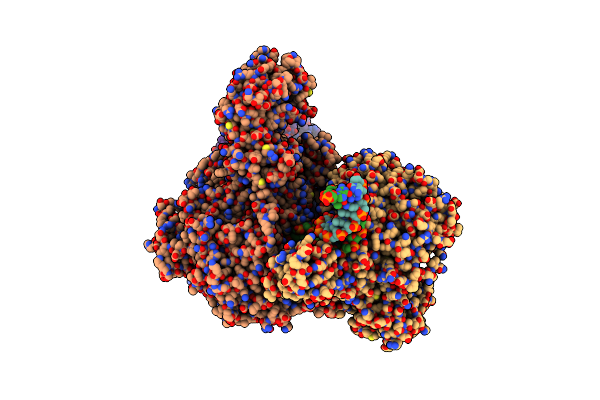 |
Organism: Vibrio cholerae
Method: ELECTRON MICROSCOPY Release Date: 2024-10-23 Classification: IMMUNE SYSTEM/DNA Ligands: MG |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.45 Å Release Date: 2024-10-16 Classification: RNA BINDING PROTEIN/RNA |
 |
Organism: Vibrio cholerae
Method: ELECTRON MICROSCOPY Release Date: 2024-09-04 Classification: IMMUNE SYSTEM/DNA |
 |
Organism: Vibrio cholerae
Method: ELECTRON MICROSCOPY Release Date: 2024-08-21 Classification: IMMUNE SYSTEM |
 |
Organism: Vibrio cholerae
Method: ELECTRON MICROSCOPY Release Date: 2024-08-21 Classification: IMMUNE SYSTEM/DNA |
 |
Organism: Maribacter polysiphoniae
Method: ELECTRON MICROSCOPY Release Date: 2023-08-23 Classification: IMMUNE SYSTEM |
 |
Structure Of Tetramerized Mapsparta Upon Guide Rna-Mediated Target Dna Binding
Organism: Maribacter polysiphoniae, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2023-08-23 Classification: IMMUNE SYSTEM Ligands: MG |
 |
Organism: Maribacter polysiphoniae, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2023-08-23 Classification: IMMUNE SYSTEM Ligands: MG |
 |
Organism: Maribacter polysiphoniae
Method: ELECTRON MICROSCOPY Release Date: 2023-08-23 Classification: IMMUNE SYSTEM Ligands: MG |
 |
Organism: Maribacter polysiphoniae
Method: ELECTRON MICROSCOPY Release Date: 2023-08-23 Classification: IMMUNE SYSTEM Ligands: MG, NAD |
 |
Organism: Maribacter polysiphoniae
Method: ELECTRON MICROSCOPY Release Date: 2023-08-23 Classification: IMMUNE SYSTEM Ligands: MG |
 |
Crystal Structure Of The Hen Egg Lysozyme-2-Oxidobenzylidene-Threoninato-Copper (Ii) Complex
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Resolution:0.92 Å Release Date: 2023-05-10 Classification: HYDROLASE Ligands: EDO, CL, JI6, NA |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.99 Å Release Date: 2022-12-28 Classification: TRANSPORT PROTEIN Ligands: 9SC, MOA |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2022-09-07 Classification: TRANSPORT PROTEIN Ligands: AJW, PO4 |
 |
Archaeal Oligopeptide Permease A (Oppa) From Thermococcus Kodakaraensis In Complex With An Endogenous Pentapeptide
Organism: Thermococcus kodakarensis (strain atcc baa-918 / jcm 12380 / kod1), Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2022-04-13 Classification: PEPTIDE BINDING PROTEIN |
 |
Crystal Structure Of Human Serum Albumin Complex With Aripiprazole And Myristic Acid
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2022-02-16 Classification: TRANSPORT PROTEIN Ligands: MYR, 9SC |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.96 Å Release Date: 2021-06-02 Classification: HYDROLASE |

