Search Count: 17
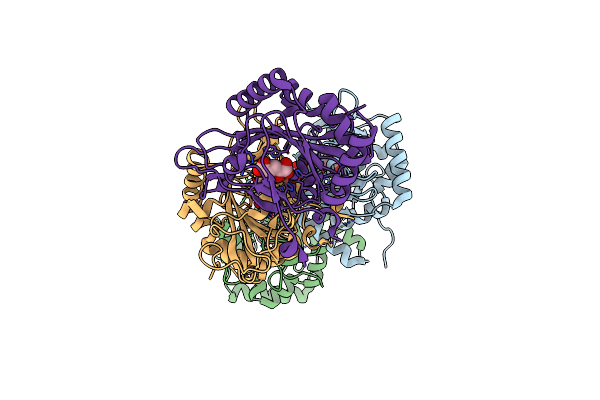 |
Crystal Structure Of The Non-Heme Alpha Ketoglutarate Dependent Epimerase Snon From Nogalamycin Biosynthesis
Organism: Streptomyces nogalater
Method: X-RAY DIFFRACTION Resolution:2.13 Å Release Date: 2016-05-11 Classification: METAL BINDING PROTEIN Ligands: AKG, FE, ACT |
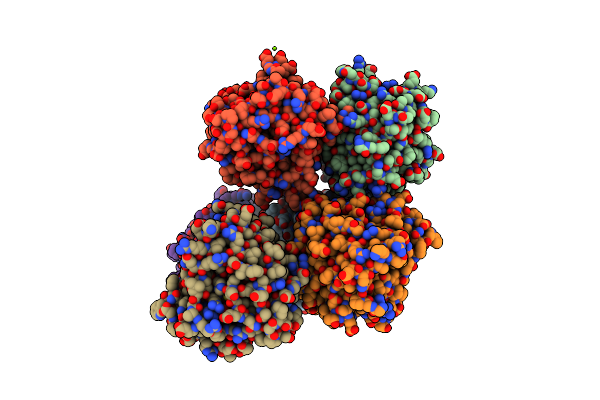 |
Crystal Structure Of Non-Heme Alpha Ketoglutarate Dependent Carbocyclase Snok From Nogalamycin Biosynthesis
Organism: Streptomyces nogalater
Method: X-RAY DIFFRACTION Resolution:2.24 Å Release Date: 2016-05-11 Classification: LYASE Ligands: FE, AKG, MG |
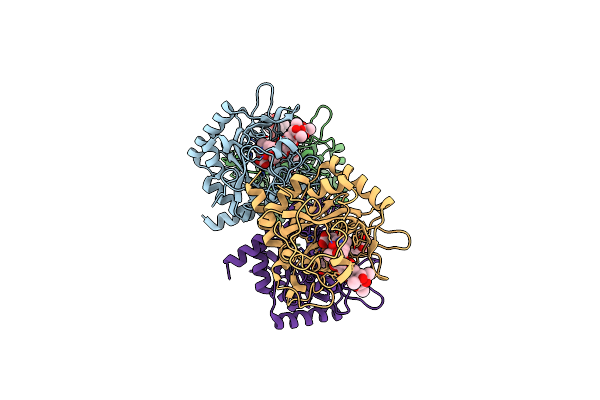 |
Crystal Structure Of The Epimerase Snon In Complex With Fe3+, Alpha Ketoglutarate And Nogalamycin Ro
Organism: Streptomyces nogalater
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2016-05-11 Classification: ISOMERASE Ligands: FE, AKG, 5R6 |
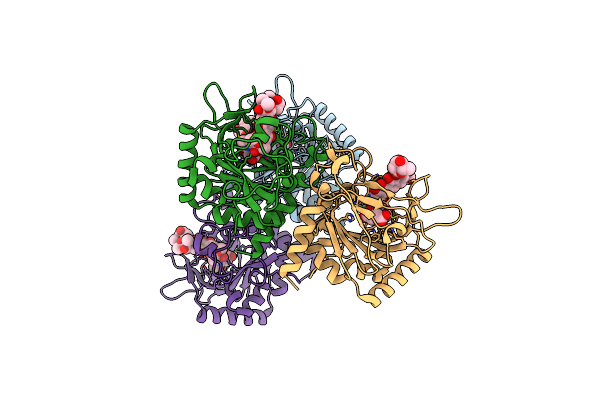 |
Crystal Structure Of The Epimerase Snon In Complex With Ni2+, Succinate And Nogalamycin Ro
Organism: Streptomyces nogalater
Method: X-RAY DIFFRACTION Resolution:2.85 Å Release Date: 2016-05-11 Classification: ISOMERASE Ligands: NI, SIN, 5R6 |
 |
Carminomycin-4-O-Methyltransferase (Dnrk) Variant (298Ser Insert) In Complex With S-Adenosyl-L-Homocysteine (Sah) And Aclacinomycin T
Organism: Streptomyces peucetius
Method: X-RAY DIFFRACTION Release Date: 2015-07-29 Classification: TRANSFERASE Ligands: SAH, 3VL, DTU, SO4 |
 |
Organism: Deinococcus radiodurans
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2015-04-29 Classification: TRANSFERASE |
 |
The Crystal Structure Of Landomycin C-6 Ketoreductase Lanv With Bound Nadp And Rabelomycin
Organism: Streptomyces cyanogenus
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2014-10-01 Classification: Oxidoreductase/antibiotic Ligands: NAP, 2V4, PEG |
 |
The Crystal Structure Of Urdamycin C-6 Ketoreductase Domain Urdmred With Bound Nadp And Rabelomycin
Organism: Streptomyces fradiae
Method: X-RAY DIFFRACTION Resolution:2.25 Å Release Date: 2014-10-01 Classification: Oxidoreductase/Antibiotic Ligands: NAP, 2V4 |
 |
Crystal Structure Of Uracil-Dna Glycosylase From Cod (Gadus Morhua) In Complex With The Proteinaceous Inhibitor Ugi
Organism: Gadus morhua, Bacillus phage pbs2
Method: X-RAY DIFFRACTION Resolution:1.93 Å Release Date: 2014-08-13 Classification: HYDROLASE/HYDROLASE INHIBITOR |
 |
The Crystal Structure Of Angucycline C-6 Ketoreductase Lanv With Bound Nadp
Organism: Streptomyces cyanogenus
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2013-07-31 Classification: OXIDOREDUCTASE Ligands: NAP, ACY, PEG |
 |
The Crystal Structure Of Angucycline C-6 Ketoreductase Lanv With Bound Nadp And 11-Deoxy-6-Oxylandomycinone
Organism: Streptomyces cyanogenus
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2013-07-31 Classification: Oxidoreductase/antibiotic Ligands: NAP, 1TJ, PEG |
 |
Organism: Streptomyces sp. cm020
Method: X-RAY DIFFRACTION Resolution:1.25 Å Release Date: 2013-01-16 Classification: HYDROLASE Ligands: MG, BO3 |
 |
Crystal Structure Of The Alnumycin P Phosphatase In Complex With Free Phosphate
Organism: Streptomyces sp. cm020
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2013-01-16 Classification: HYDROLASE Ligands: MG, BO3, PO4 |
 |
Organism: Streptomyces sp. cm020
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2013-01-16 Classification: LIGASE Ligands: CA, SO4, CL, PEG |
 |
Crystal Structure Of The Prealnumycin C-Glycosynthase Alna In Complex With Ribulose 5-Phosphate
Organism: Streptomyces sp. cm020
Method: X-RAY DIFFRACTION Resolution:3.15 Å Release Date: 2013-01-16 Classification: LIGASE Ligands: CA, 5RP, EPE, CL |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2011-10-12 Classification: HYDROLASE Ligands: IMD |
 |
Organism: Vibrio cholerae
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2008-02-12 Classification: HYDROLASE Ligands: CL, MG |

