Search Count: 23
 |
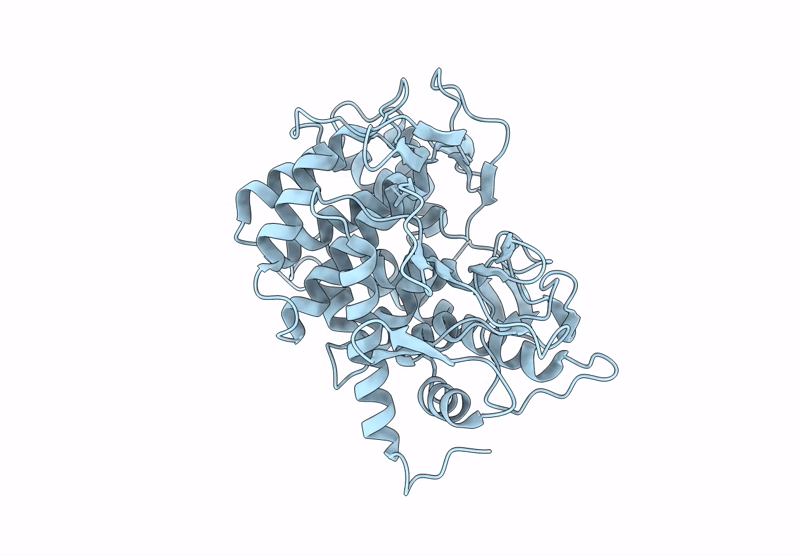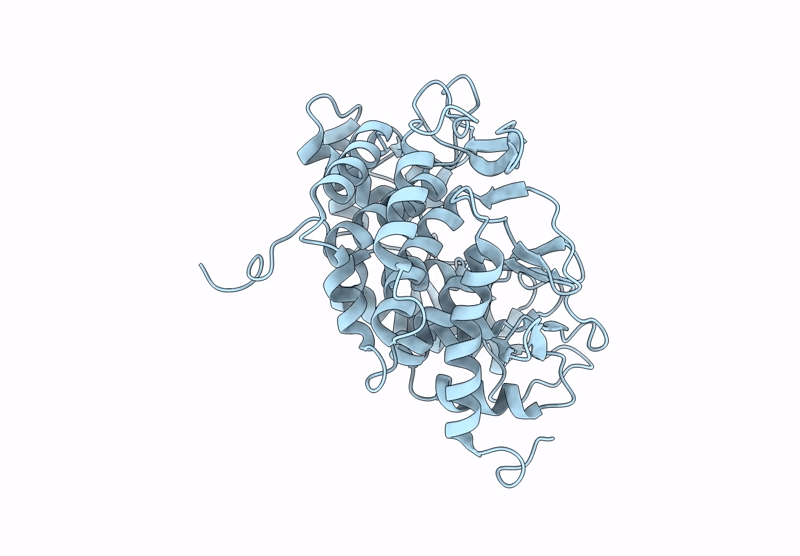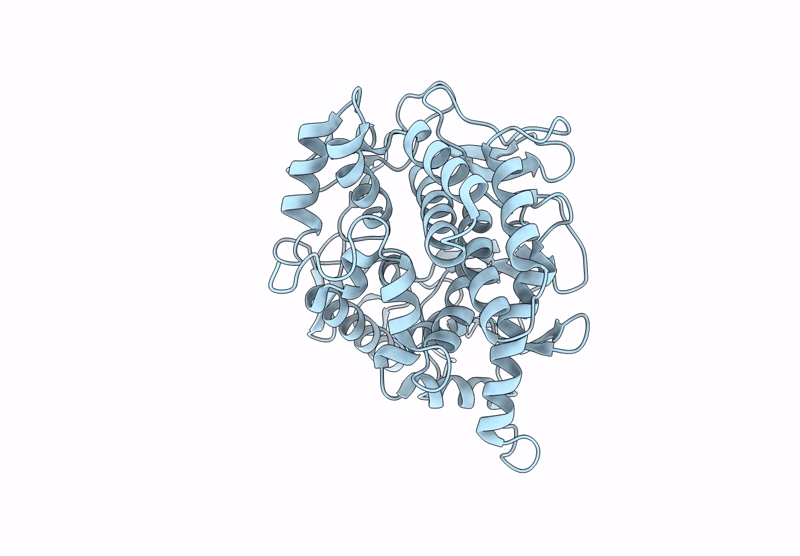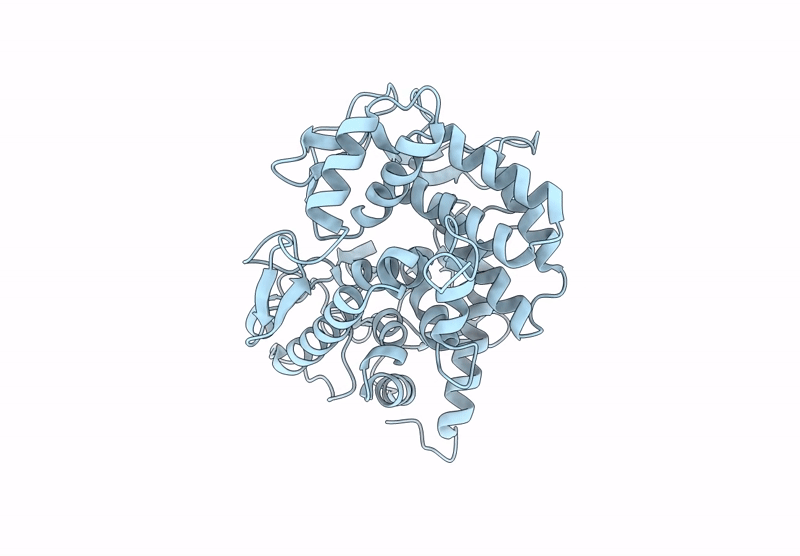
Structure Of Beta-1,2-Glucanase From Endozoicomonas Elysicola (Eesgl1, Ligand-Free)
Organism: Endozoicomonas elysicola dsm 22380
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2025-01-22 Classification: HYDROLASE |
 |
Structure Of Beta-1,2-Glucanase From Photobacterium Gaetbulicola (Pgsgl3, Ligand-Free)
Organism: Photobacterium gaetbulicola gung47
Method: X-RAY DIFFRACTION Resolution:1.20 Å Release Date: 2025-01-22 Classification: HYDROLASE Ligands: CL |
 |
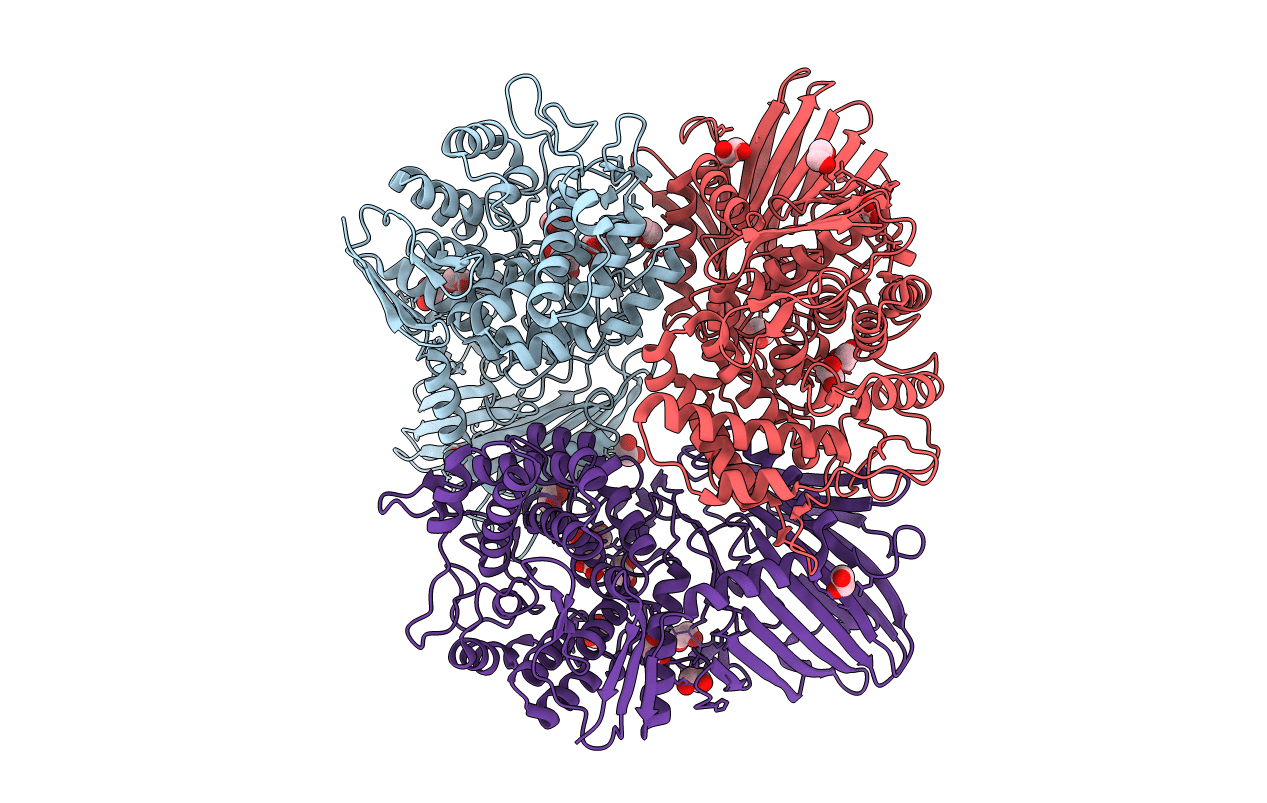
Structure Of Beta-1,2-Glucanase From Xanthomonas Campestris Pv. Campestris (Beta-1,2-Glucoheptasaccharide Complex)-E239Q Mutant
Organism: Xanthomonas campestris pv. campestris
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2025-01-22 Classification: HYDROLASE |
 |
Crystal Structure Of Gh65 Alpha-1,2-Glucosidase From Flavobacterium Johnsoniae
Organism: Flavobacterium johnsoniae (strain atcc 17061 / dsm 2064 / jcm 8514 / nbrc 14942 / ncimb 11054 / uw101)
Method: X-RAY DIFFRACTION Resolution:1.54 Å Release Date: 2021-11-10 Classification: HYDROLASE Ligands: EDO |
 |
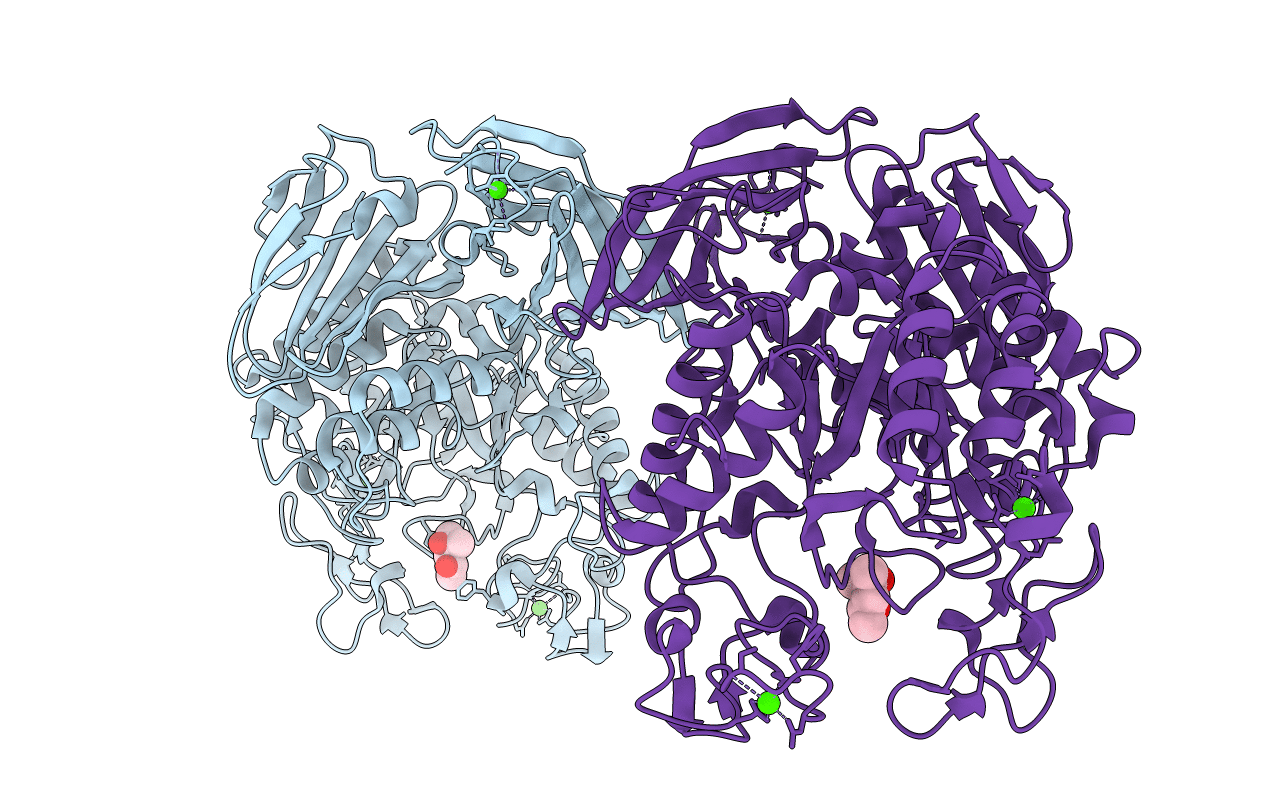
Crystal Structure Of Gh65 Alpha-1,2-Glucosidase From Flavobacterium Johnsoniae In Complex With Glucose
Organism: Flavobacterium johnsoniae (strain atcc 17061 / dsm 2064 / jcm 8514 / nbrc 14942 / ncimb 11054 / uw101)
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2021-11-10 Classification: HYDROLASE Ligands: BGC |
 |
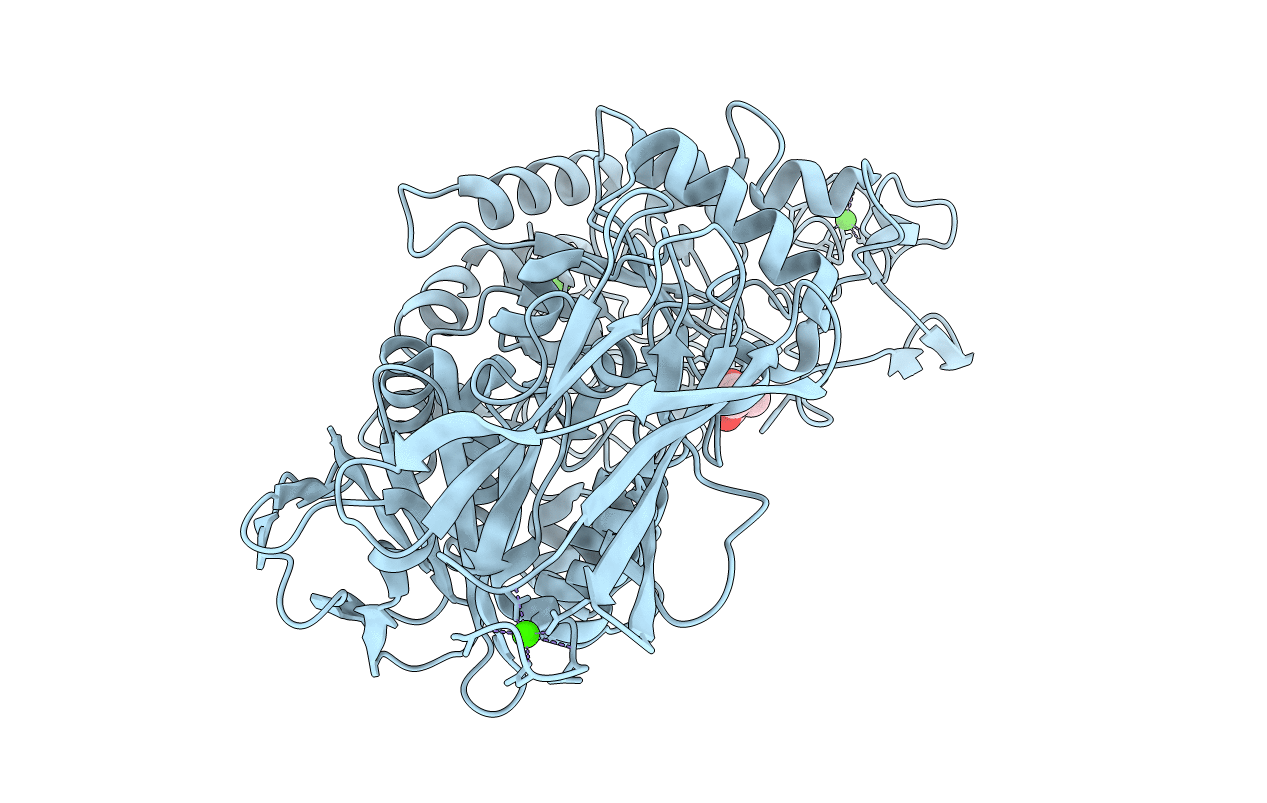
Thermoactinomyces Vulgaris R-47 Alpha-Amylase I (Tva I) Mutant A357V/Q359N/Y360E (Aqy/Vne)
Organism: Thermoactinomyces vulgaris
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2018-02-21 Classification: HYDROLASE Ligands: CA, MPD |
 |
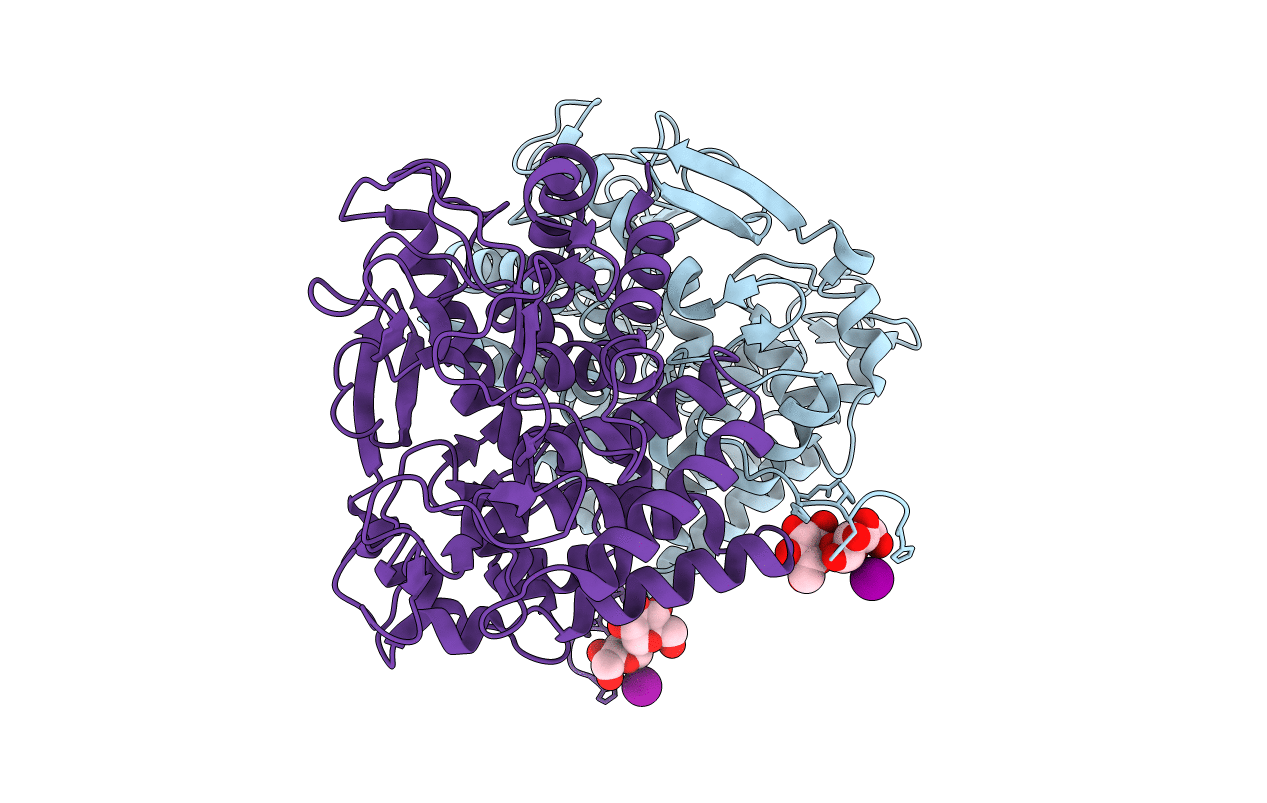
Thermoactinomyces Vulgaris R-47 Alpha-Amylase I (Tva I) 11 Residues (From A363 To N373) Deletion Mutant (Del11)
Organism: Thermoactinomyces vulgaris
Method: X-RAY DIFFRACTION Resolution:1.37 Å Release Date: 2018-02-21 Classification: HYDROLASE Ligands: CA, MPD |
 |
Organism: Chitinophaga pinensis (strain atcc 43595 / dsm 2588 / ncib 11800 / uqm 2034)
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2017-03-15 Classification: HYDROLASE Ligands: IOD |
 |
Endo-Beta-1,2-Glucanase From Chitinophaga Pinensis - Sophorotriose And Glucose Complex
Organism: Chitinophaga pinensis (strain atcc 43595 / dsm 2588 / ncib 11800 / uqm 2034)
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2017-03-15 Classification: HYDROLASE Ligands: PO4, K, BGC, PG4, PEG, CL, 1PE |
 |
Crystal Structure Of 1,2-Beta-Oligoglucan Phosphorylase From Lachnoclostridium Phytofermentans
Organism: Clostridium phytofermentans isdg
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2017-03-01 Classification: TRANSFERASE Ligands: GOL, CA, PEG |
 |
Crystal Structure Of 1,2-Beta-Oligoglucan Phosphorylase From Lachnoclostridium Phytofermentans In Complex With Sophorose
Organism: Clostridium phytofermentans isdg
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2017-03-01 Classification: TRANSFERASE Ligands: GOL, CA |
 |
Crystal Structure Of 1,2-Beta-Oligoglucan Phosphorylase From Lachnoclostridium Phytofermentans In Complex With Sophorose, Isofagomine, Sulfate Ion
Organism: Clostridium phytofermentans isdg
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2017-03-01 Classification: TRANSFERASE Ligands: IFM, SO4 |
 |
Crystal Structure Of 1,2-Beta-Oligoglucan Phosphorylase From Lachnoclostridium Phytofermentans In Complex With Alpha-D-Glucose-1-Phosphate
Organism: Clostridium phytofermentans isdg
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2017-03-01 Classification: TRANSFERASE Ligands: G1P, GLC |
 |
Organism: Listeria innocua clip11262
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2015-12-02 Classification: TRANSFERASE Ligands: SO4, GOL, MES |
 |
Organism: Listeria innocua clip11262
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2015-12-02 Classification: TRANSFERASE Ligands: MAN, SO4, MES |
 |
Beta-1,2-Mannobiose Phosphorylase From Listeria Innocua - Beta-1,2-Mannobiose Complex
Organism: Listeria innocua clip11262
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2015-12-02 Classification: TRANSFERASE Ligands: SO4, GOL, MES |
 |
Beta-1,2-Mannobiose Phosphorylase From Listeria Innocua - Beta-1,2-Mannotriose Complex
Organism: Listeria innocua clip11262
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2015-12-02 Classification: TRANSFERASE Ligands: SO4, MES, GOL |
 |
Organism: Saccharophagus degradans 2-40
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2015-06-10 Classification: TRANSFERASE Ligands: SO4, GOL, CL |
 |
Organism: Saccharophagus degradans 2-40
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2015-06-10 Classification: TRANSFERASE Ligands: CEZ, SO4, GOL, CL |
 |
Organism: Saccharophagus degradans 2-40
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2015-06-10 Classification: TRANSFERASE Ligands: GCO, GOL, LGC, SO4, CL |

