Search Count: 71
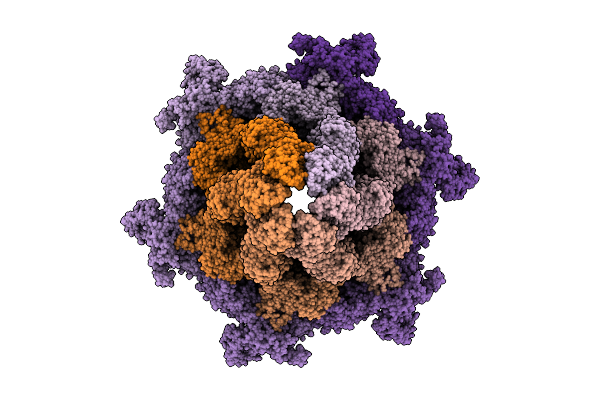 |
Cryo-Em Structure Of The Plpvc1 Baseplate, 6-Fold Symmetrized (C6), In Extended State
Organism: Photorhabdus luminescens
Method: ELECTRON MICROSCOPY Release Date: 2025-10-29 Classification: VIRUS LIKE PARTICLE |
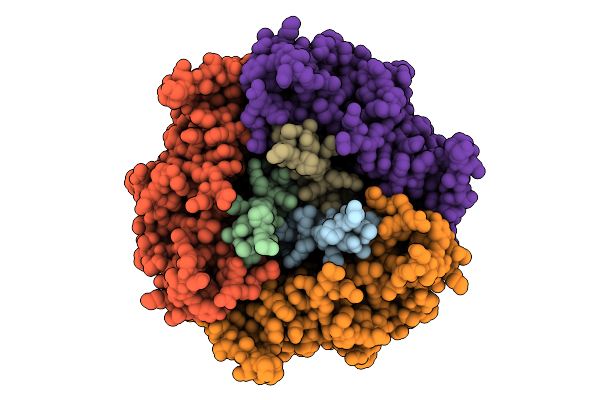 |
Cryo-Em Structure Of The Plpvc1 Central Spike, 3-Fold Symmetrized (C3), In Extended State
Organism: Photorhabdus luminescens
Method: ELECTRON MICROSCOPY Release Date: 2025-10-29 Classification: VIRUS LIKE PARTICLE |
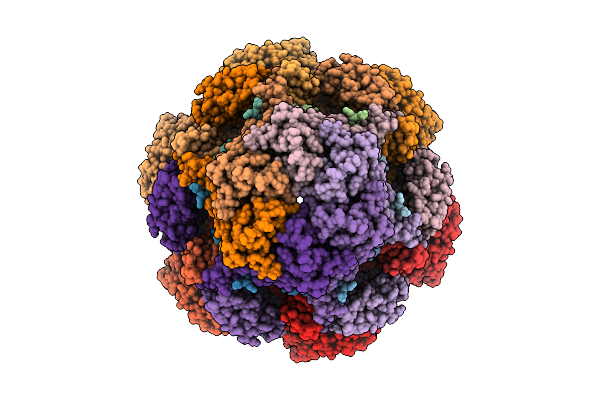 |
Cryo-Em Structure Of The Plpvc1 Cap, 6-Fold Symmetrized (C6), In Extended State
Organism: Photorhabdus luminescens
Method: ELECTRON MICROSCOPY Release Date: 2025-10-29 Classification: VIRUS LIKE PARTICLE |
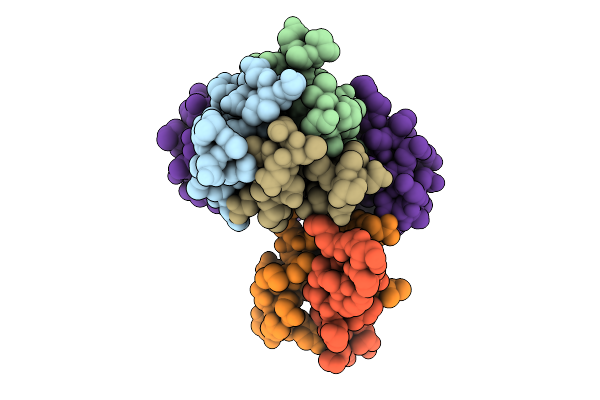 |
Organism: Photorhabdus luminescens
Method: ELECTRON MICROSCOPY Release Date: 2025-10-29 Classification: VIRUS LIKE PARTICLE |
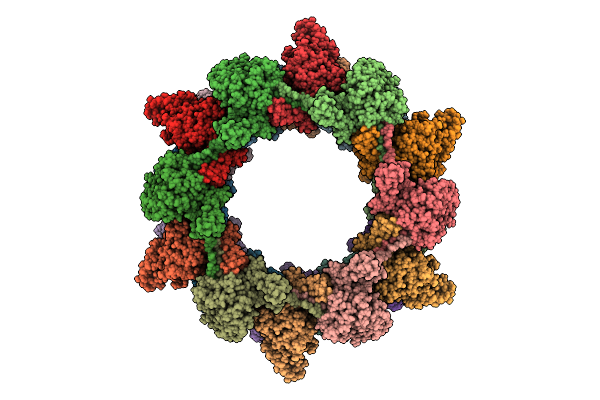 |
Cryo-Em Structure Of The Plpvc1 Sheath, 6-Fold Symmetrized (C6), In Contracted State
Organism: Photorhabdus luminescens
Method: ELECTRON MICROSCOPY Release Date: 2025-10-29 Classification: VIRUS LIKE PARTICLE |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2024-12-18 Classification: IMMUNE SYSTEM Ligands: GOL, PEG, NAG |
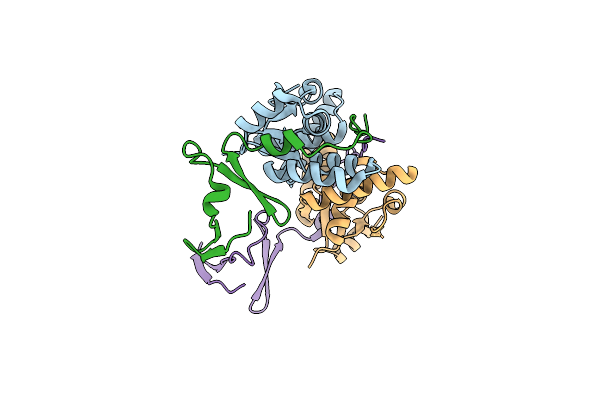 |
Organism: Escherichia coli kly
Method: X-RAY DIFFRACTION Resolution:2.86 Å Release Date: 2024-12-18 Classification: TOXIN |
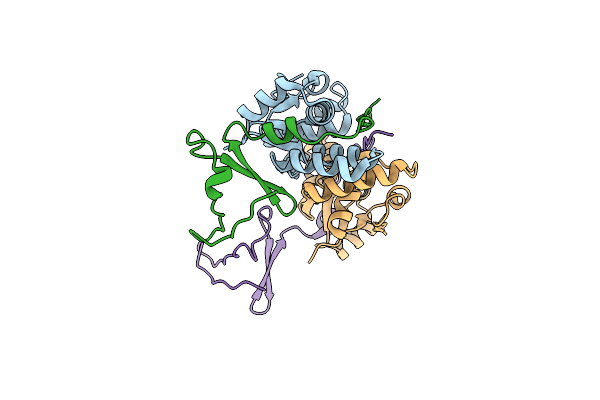 |
Crystal Structure Of The E. Coli F-Plasmid Vapbc Toxin-Antitoxin Complex (Vapb T3N, A13P, L16R)
Organism: Escherichia coli kly
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2024-12-18 Classification: TOXIN |
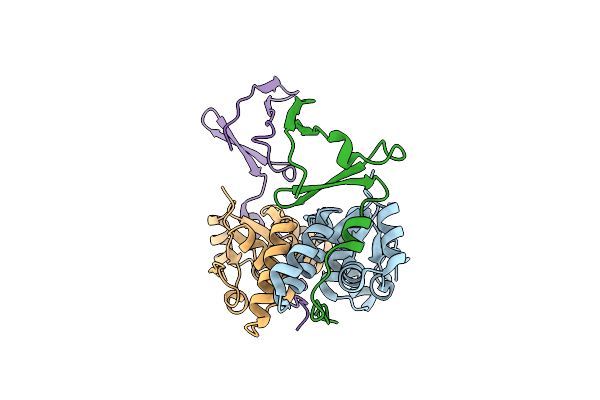 |
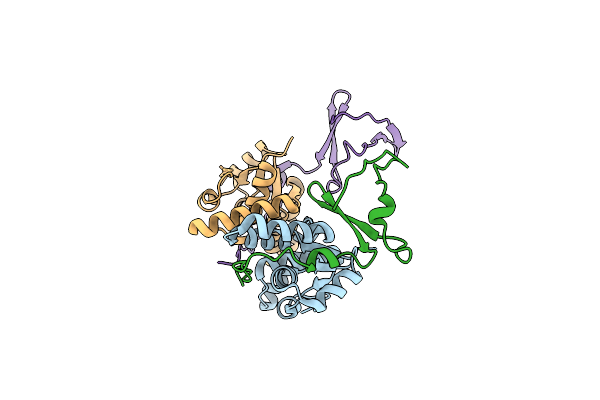
Crystal Structure Of The E. Coli F-Plasmid Vapbc Toxin-Antitoxin Complex (Vapb T3N)
Organism: Escherichia coli kly
Method: X-RAY DIFFRACTION Resolution:2.65 Å Release Date: 2024-12-18 Classification: TOXIN |
 |
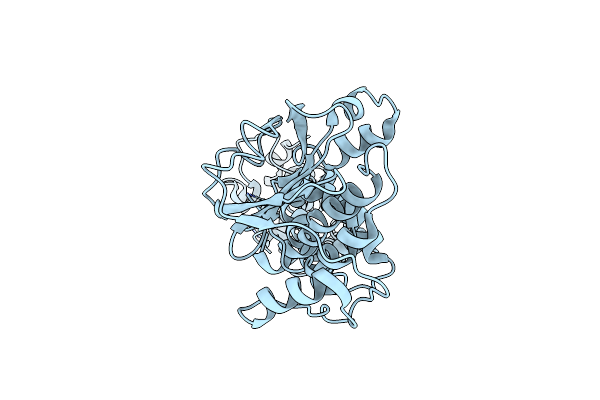
Crystal Structure Of The E. Coli F-Plasmid Vapbc Toxin-Antitoxin Complex (Vapb V5E)
Organism: Escherichia coli kly
Method: X-RAY DIFFRACTION Resolution:3.15 Å Release Date: 2024-12-18 Classification: TOXIN |
 |
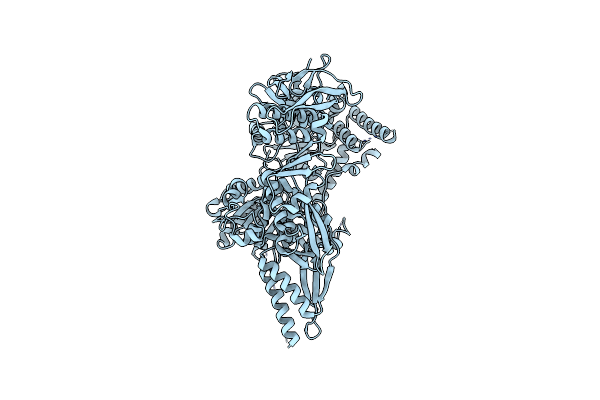
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2024-12-04 Classification: ANTIVIRAL PROTEIN |
 |
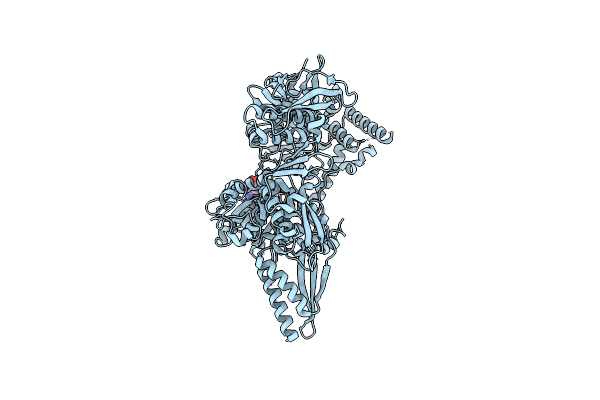
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2024-11-06 Classification: ANTIVIRAL PROTEIN |
 |
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2024-11-06 Classification: ANTIVIRAL PROTEIN Ligands: AGS |
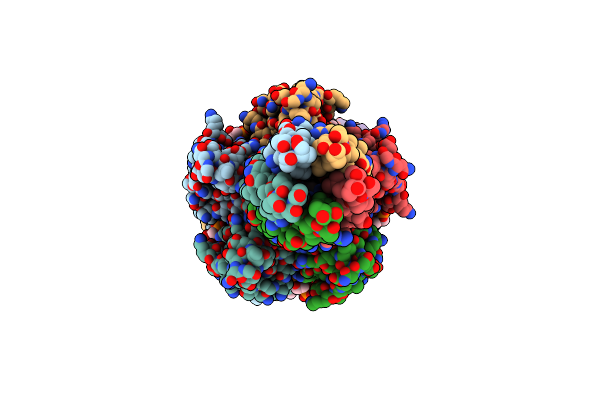 |
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2024-11-06 Classification: ANTIVIRAL PROTEIN Ligands: CDL, CA, PEE, PLM |
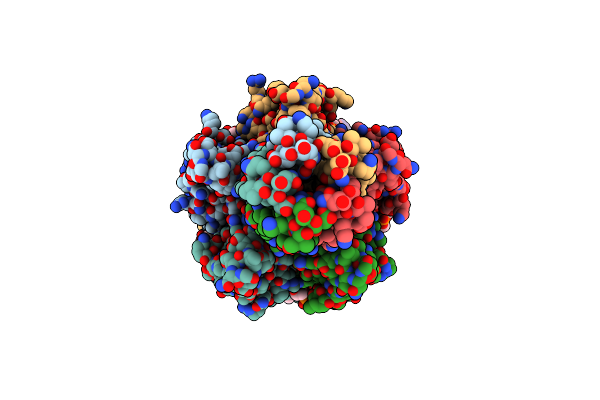 |
Zorya Anti-Bacteriophage Defense System Zorab Zora E86A_E89A, Calcium Binding Site Mutation
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2024-11-06 Classification: ANTIVIRAL PROTEIN Ligands: CDL, PEE |
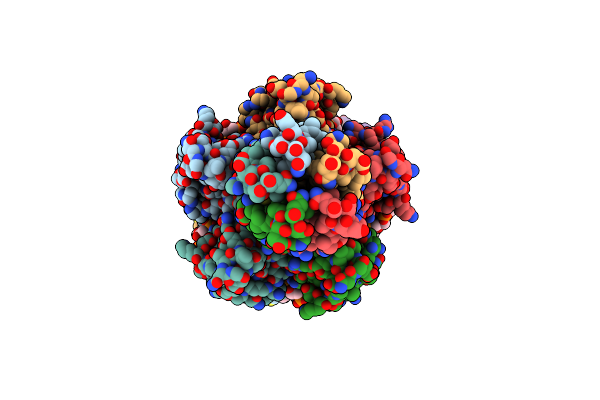 |
Zorya Anti-Bacteriophage Defense System Zorab, Zora Delta_359-592, Zora Tail Middle Deletion.
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2024-11-06 Classification: ANTIVIRAL PROTEIN Ligands: CDL, PEE, CA |
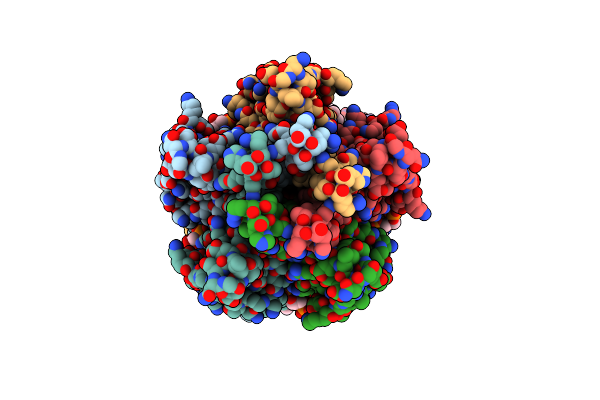 |
Zorya Anti-Bacteriophage Defense System Zorab, Zora Delta_435-729, Zora Tail Tip Deletion.
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2024-11-06 Classification: ANTIVIRAL PROTEIN Ligands: CDL, CA, PEE |
 |
Organism: Homo sapiens, Lama glama
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2024-07-17 Classification: MEMBRANE PROTEIN Ligands: A1AE2, AV0 |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-06-05 Classification: SUGAR BINDING PROTEIN |
 |
Retron-Eco1 Filament With Adp-Ribosylated Effector (Local Map With 1 Segment)
Organism: Escherichia coli bl21(de3)
Method: ELECTRON MICROSCOPY Release Date: 2024-06-05 Classification: IMMUNE SYSTEM Ligands: MG, AR6 |

