Search Count: 40
 |
Organism: Haemophilus influenzae
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2014-12-31 Classification: LYASE Ligands: ZN, SO4 |
 |
Organism: Haemophilus influenzae
Method: X-RAY DIFFRACTION Resolution:2.49 Å Release Date: 2014-12-31 Classification: LYASE Ligands: ZN, BCT, K |
 |
Organism: Haemophilus influenzae
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2014-12-31 Classification: LYASE Ligands: ZN, PO4 |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 1997-03-12 Classification: IRON TRANSPORT Ligands: FE, CO3 |
 |
Organism: Enterobacteria phage t4
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 1993-10-31 Classification: HYDROLASE(O-GLYCOSYL) Ligands: CL, BME |
 |
Organism: Enterobacteria phage t4
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 1993-10-31 Classification: HYDROLASE(O-GLYCOSYL) |
 |
Hydrophobic Core Repacking And Aromatic-Aromatic Interaction In The Thermostable Mutant Of T4 Lysozyme Ser 117 (Right Arrow) Phe
Organism: Enterobacteria phage t4
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 1993-07-15 Classification: HYDROLASE (O-GLYCOSYL) Ligands: PO4, CL |
 |
Comparison Of The Crystal Structure Of Bacteriophage T4 Lysozyme At Low, Medium, And High Ionic Strengths
Organism: Enterobacteria phage t4
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 1992-07-15 Classification: HYDROLASE (O-GLYCOSYL) Ligands: CL, BME |
 |
Comparison Of The Crystal Structure Of Bacteriophage T4 Lysozyme At Low, Medium, And High Ionic Strengths
Organism: Enterobacteria phage t4
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 1992-07-15 Classification: HYDROLASE (O-GLYCOSYL) Ligands: CL, BME |
 |
Comparison Of The Crystal Structure Of Bacteriophage T4 Lysozyme At Low, Medium, And High Ionic Strengths
Organism: Enterobacteria phage t4
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 1992-07-15 Classification: HYDROLASE (O-GLYCOSYL) Ligands: CL, BME |
 |
Comparison Of The Crystal Structure Of Bacteriophage T4 Lysozyme At Low, Medium, And High Ionic Strengths
Organism: Enterobacteria phage t4
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 1992-07-15 Classification: HYDROLASE (O-GLYCOSYL) Ligands: CL |
 |
Contributions Of Engineered Surface Salt Bridges To The Stability Of T4 Lysozyme Determined By Directed Mutagenesis
Organism: Enterobacteria phage t4
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 1991-10-15 Classification: HYDROLASE (O-GLYCOSYL) |
 |
Contributions Of Engineered Surface Salt Bridges To The Stability Of T4 Lysozyme Determined By Directed Mutagenesis
Organism: Enterobacteria phage t4
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 1991-10-15 Classification: HYDROLASE (O-GLYCOSYL) |
 |
Contributions Of Engineered Surface Salt Bridges To The Stability Of T4 Lysozyme Determined By Directed Mutagenesis
Organism: Enterobacteria phage t4
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 1991-10-15 Classification: HYDROLASE (O-GLYCOSYL) |
 |
Contributions Of Engineered Surface Salt Bridges To The Stability Of T4 Lysozyme Determined By Directed Mutagenesis
Organism: Enterobacteria phage t4
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 1991-10-15 Classification: HYDROLASE (O-GLYCOSYL) |
 |
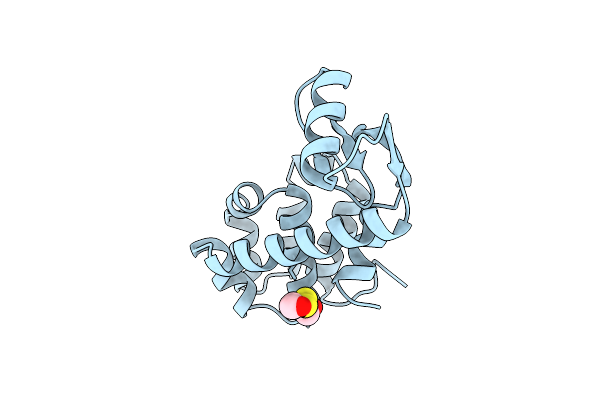
Contributions Of Engineered Surface Salt Bridges To The Stability Of T4 Lysozyme Determined By Directed Mutagenesis
Organism: Enterobacteria phage t4
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 1991-10-15 Classification: HYDROLASE (O-GLYCOSYL) |
 |
Analysis Of The Interaction Between Charged Side Chains And The Alpha-Helix Dipole Using Designed Thermostable Mutants Of Phage T4 Lysozyme
Organism: Enterobacteria phage t4
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 1991-10-15 Classification: HYDROLASE (O-GLYCOSYL) Ligands: CL, BME |
 |
Analysis Of The Interaction Between Charged Side Chains And The Alpha-Helix Dipole Using Designed Thermostable Mutants Of Phage T4 Lysozyme
Organism: Enterobacteria phage t4
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 1991-10-15 Classification: HYDROLASE (O-GLYCOSYL) Ligands: BME |
 |
Analysis Of The Interaction Between Charged Side Chains And The Alpha-Helix Dipole Using Designed Thermostable Mutants Of Phage T4 Lysozyme
Organism: Enterobacteria phage t4
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 1991-10-15 Classification: HYDROLASE (O-GLYCOSYL) Ligands: CL, BME |
 |
Analysis Of The Interaction Between Charged Side Chains And The Alpha-Helix Dipole Using Designed Thermostable Mutants Of Phage T4 Lysozyme
Organism: Enterobacteria phage t4
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 1991-10-15 Classification: HYDROLASE (O-GLYCOSYL) Ligands: BME |

