Search Count: 20
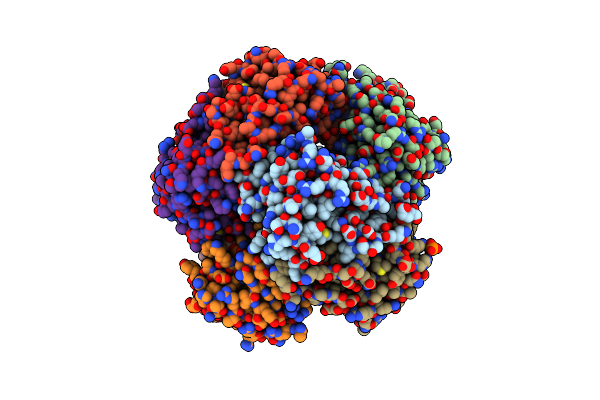 |
Crystal Structure Of Anubix (Pada1) In Complex With Fmn And Dimethylallyl Pyrophosphate
Organism: Aspergillus niger
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2019-06-05 Classification: TRANSFERASE Ligands: DMA, FMN, PO4, PEG |
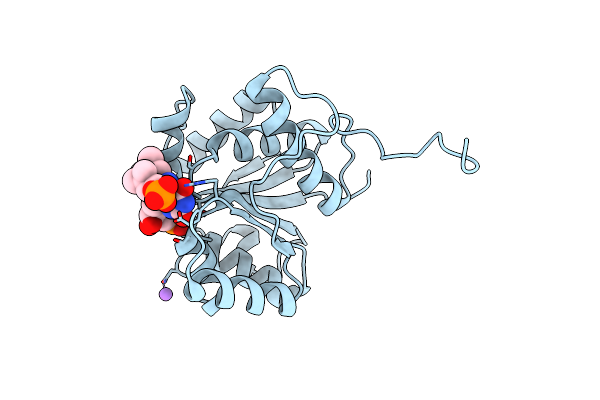 |
Crystal Structure Of Ubix In Complex With Reduced Fmn And Isopentyl Monophosphate
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:1.57 Å Release Date: 2019-06-05 Classification: TRANSFERASE |
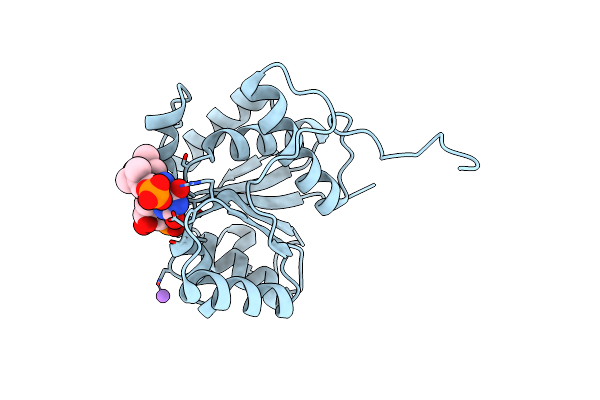 |
Crystal Structure Of F181Q Ubix In Complex With Fmn And Dimethylallyl Monophosphate
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:1.99 Å Release Date: 2019-06-05 Classification: TRANSFERASE Ligands: 4LR, FNR, NA |
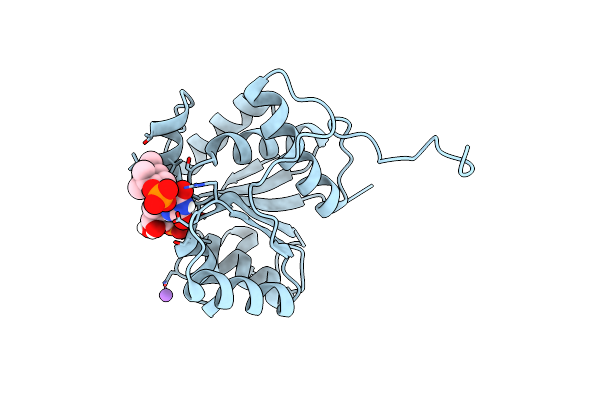 |
Crystal Structure Of F181Q Ubix In Complex With An Oxidised N5-C1' Adduct Derived From Dmap
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:1.77 Å Release Date: 2019-06-05 Classification: TRANSFERASE Ligands: 4LS, NA, PO4 |
 |
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:1.78 Å Release Date: 2019-06-05 Classification: TRANSFERASE Ligands: 4LU, PO4, NA |
 |
Crystal Structure Of F181H Ubix In Complex With Fmn And Dimethylallyl Monophosphate
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:1.56 Å Release Date: 2019-06-05 Classification: TRANSFERASE Ligands: 4LR, FNR, NA, SO4 |
 |
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:2.39 Å Release Date: 2019-06-05 Classification: TRANSFERASE Ligands: HZZ, FMN, ACT, NA, HJN, PO4 |
 |
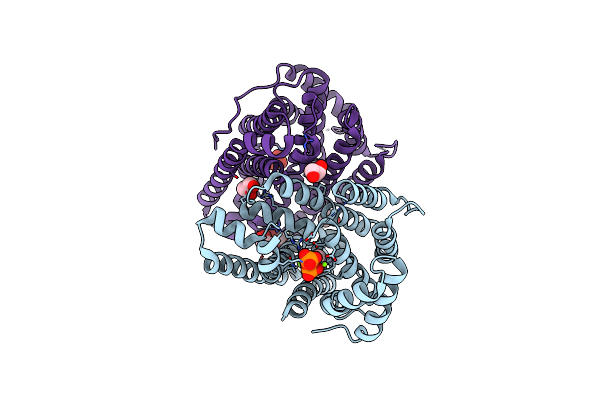
Crystal Structure Of Linalool/Nerolidol Synthase From Streptomyces Clavuligerus
Organism: Streptomyces clavuligerus
Method: X-RAY DIFFRACTION Resolution:2.38 Å Release Date: 2017-09-20 Classification: LIGASE Ligands: GOL |
 |
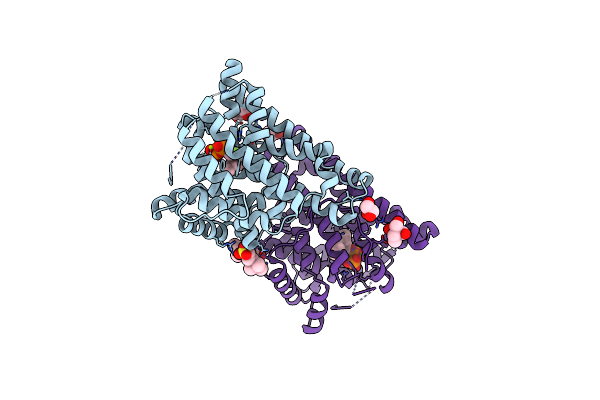
Crystal Structure Of Linalool/Nerolidol Synthase From Streptomyces Clavuligerus In Complex With 2-Fluorogeranyl Diphosphate
Organism: Streptomyces clavuligerus
Method: X-RAY DIFFRACTION Resolution:1.82 Å Release Date: 2017-09-20 Classification: LIGASE Ligands: MG, 0FV, CL, GOL, PO4 |
 |
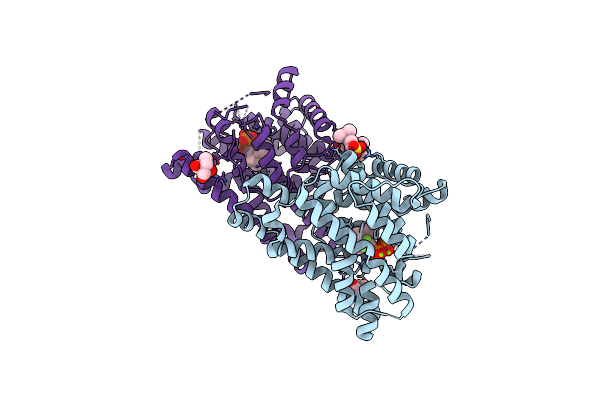
Crystal Structure Of 1,8-Cineole Synthase From Streptomyces Clavuligerus In Complex With 2-Fluoroneryl Diphosphate
Organism: Streptomyces clavuligerus
Method: X-RAY DIFFRACTION Resolution:1.63 Å Release Date: 2017-09-20 Classification: LYASE Ligands: LA6, MG, 9D2, NDS, GOL |
 |
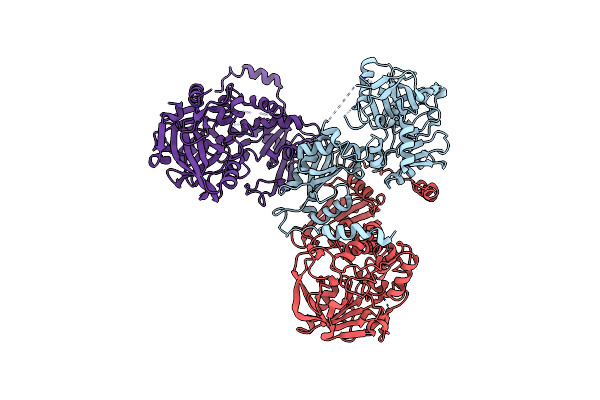
Crystal Structure Of 1,8-Cineole Synthase From Streptomyces Clavuligerus In Complex With 2-Fluoroneryl Diphosphate And 2-Fluorogeranyl Diphosphate
Organism: Streptomyces clavuligerus
Method: X-RAY DIFFRACTION Resolution:1.51 Å Release Date: 2017-09-20 Classification: LYASE Ligands: 0FV, LA6, MG, 9D2, NDS |
 |
Organism: Escherichia coli o6:h1 (strain cft073 / atcc 700928 / upec)
Method: X-RAY DIFFRACTION Resolution:3.15 Å Release Date: 2017-01-11 Classification: LYASE |
 |
Organism: Escherichia coli o6:h1 (strain cft073 / atcc 700928 / upec)
Method: X-RAY DIFFRACTION Resolution:2.75 Å Release Date: 2017-01-11 Classification: LYASE Ligands: PG4 |
 |
Crystal Structure Of N-Terminally Tagged Ubid From E. Coli Reconstituted With Prfmn Cofactor
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2017-01-11 Classification: LYASE Ligands: MN, 4LU, NA, SO4 |
 |
Crystal Structure Of N-Terminally Tagged Ubid From E. Coli Reconstituted With Prfmn Cofactor
Organism: Escherichia coli o6:h1 (strain cft073 / atcc 700928 / upec)
Method: X-RAY DIFFRACTION Resolution:2.62 Å Release Date: 2017-01-11 Classification: LYASE Ligands: MN, 7D9, NA |
 |
Isopiperitenone Reductase From Mentha Piperita In Complex With Nadp And Beta-Cyclocitral
Organism: Mentha piperita
Method: X-RAY DIFFRACTION Resolution:1.41 Å Release Date: 2016-08-31 Classification: OXIDOREDUCTASE Ligands: 6KX, NAP |
 |
Organism: Mentha piperita
Method: X-RAY DIFFRACTION Resolution:2.66 Å Release Date: 2016-08-31 Classification: OXIDOREDUCTASE |
 |
Organism: Mentha piperita
Method: X-RAY DIFFRACTION Resolution:2.24 Å Release Date: 2016-08-31 Classification: OXIDOREDUCTASE Ligands: NAP |
 |
Organism: Mentha piperita
Method: X-RAY DIFFRACTION Resolution:1.71 Å Release Date: 2016-08-31 Classification: OXIDOREDUCTASE Ligands: NAP, MPD |
 |
Isopiperitenone Reductase From Mentha Piperita In Complex With Isopiperitenone And Nadp
Organism: Mentha piperita
Method: X-RAY DIFFRACTION Resolution:1.30 Å Release Date: 2016-08-31 Classification: OXIDOREDUCTASE Ligands: NAP, IT9 |

