Search Count: 307
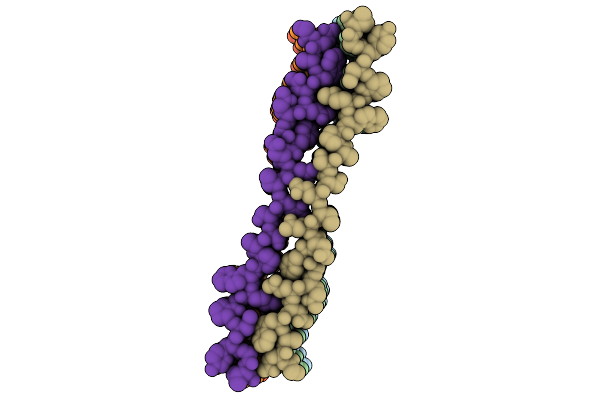 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-12-03 Classification: PROTEIN FIBRIL |
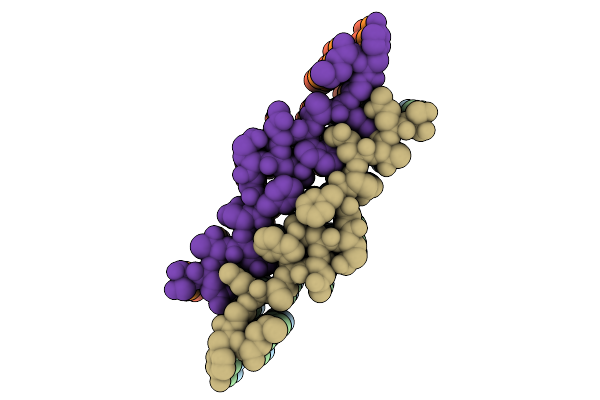 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-12-03 Classification: PROTEIN FIBRIL |
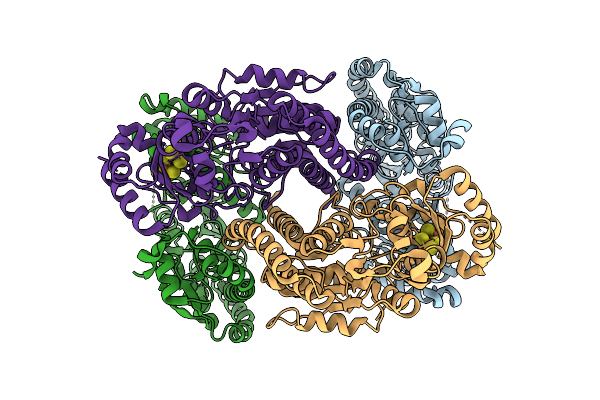 |
Rhodospirillum Rubrum Nitrogenase-Like Methylthio-Alkane Reductase Complex With An Oxidized P-Cluster
Organism: Rhodospirillum rubrum atcc 11170
Method: ELECTRON MICROSCOPY Release Date: 2025-11-05 Classification: OXIDOREDUCTASE Ligands: CLF |
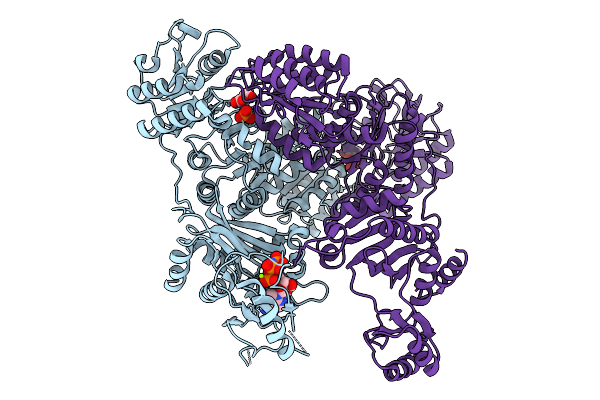 |
3-Hydroxypropionyl-Coa Synthetase (Adp-Forming) From Nitrosopumilus Maritimus.
Organism: Nitrosopumilus maritimus scm1
Method: X-RAY DIFFRACTION Release Date: 2025-10-01 Classification: LIGASE Ligands: ANP, 3OH, PO4, MG |
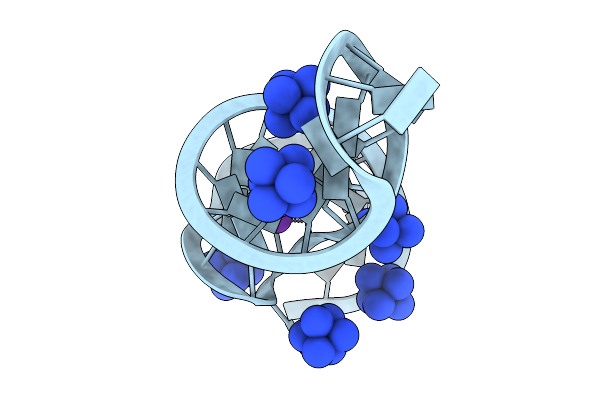 |
Crystal Structure Of Theophylline Aptamer Obtained In The Presence Of Caffeine
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-09-03 Classification: RNA Ligands: NCO, K |
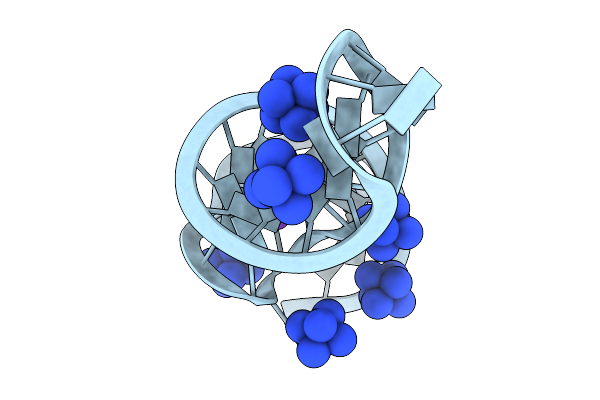 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-09-03 Classification: RNA Ligands: NCO, K |
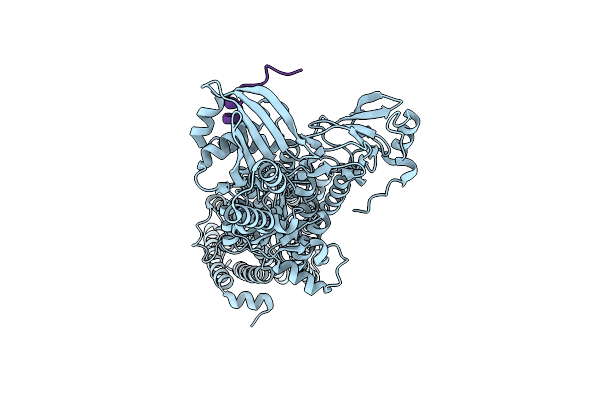 |
Cryo-Em Structure Of The Pma1 With Ordered N-Terminal Extension In The Activated State
Organism: Saccharomyces cerevisiae (strain atcc 204508 / s288c)
Method: ELECTRON MICROSCOPY Release Date: 2025-06-04 Classification: PROTON TRANSPORT |
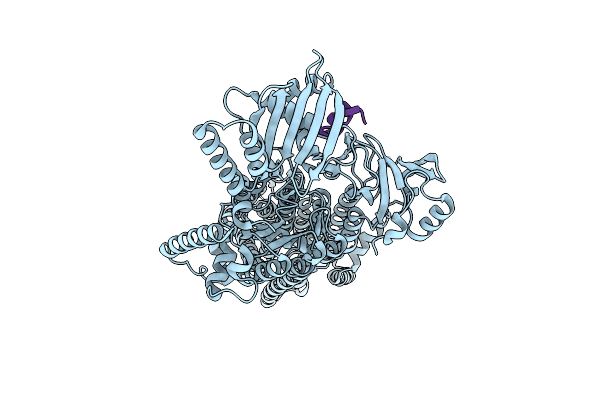 |
Cryo-Em Structure Of The Pma1 With Ordered N-Terminal Extension In The Autoinhibited State
Organism: Saccharomyces cerevisiae (strain atcc 204508 / s288c)
Method: ELECTRON MICROSCOPY Release Date: 2025-06-04 Classification: PROTON TRANSPORT |
 |
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Release Date: 2025-05-28 Classification: HYDROLASE Ligands: GD, DO3, NO3, CL, NA |
 |
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Release Date: 2025-05-28 Classification: HYDROLASE Ligands: GD, DO3, NO3, CL, NA |
 |
Organism: Modestobacter roseus
Method: X-RAY DIFFRACTION Resolution:2.87 Å Release Date: 2025-04-23 Classification: OXIDOREDUCTASE Ligands: NAD, ZN |
 |
Organism: Modestobacter roseus
Method: X-RAY DIFFRACTION Resolution:2.59 Å Release Date: 2025-04-23 Classification: OXIDOREDUCTASE Ligands: NAD, ZN |
 |
Organism: Takifugu rubripes
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2025-04-02 Classification: IMMUNE SYSTEM Ligands: GLC, BGC, CA |
 |
Organism: Takifugu rubripes
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2025-04-02 Classification: IMMUNE SYSTEM Ligands: GOL, CA |
 |
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2025-03-19 Classification: RIBOSOME Ligands: MG, K, SPD, PUT, ZN |
 |
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2025-03-19 Classification: RIBOSOME Ligands: MG, K, SPD, PUT, ZN |
 |
X-Ray Structure Of The Drug Binding Domain Of Alba In Complex With The Kmr-14-14 Compound Of The Pyrrolobenzodiazepines Class
Organism: Klebsiella oxytoca
Method: X-RAY DIFFRACTION Resolution:2.17 Å Release Date: 2025-01-15 Classification: DNA BINDING PROTEIN Ligands: A1H1R, A1H1Q, DTV |
 |
Organism: Grindelia hirsutula
Method: X-RAY DIFFRACTION Resolution:2.12 Å Release Date: 2024-11-06 Classification: PLANT PROTEIN |
 |
Crystal Structure Of 2-Hydroxacyl-Coa Lyase/Synthase Apbhacs From Alphaproteobacteria Bacterium In The Complex With Thdp, L-Lactyl-Coa, And Adp
Organism: Alphaproteobacteria bacterium
Method: X-RAY DIFFRACTION Resolution:2.05 Å Release Date: 2024-10-02 Classification: LYASE Ligands: 8FL, UQ3, ADP, ACE, MG, GOL |
 |
Crystal Structure Of 2-Hydroxacyl-Coa Lyase/Synthase Apbhacs From Alphaproteobacteria Bacterium In The Complex With Thdp, D-Lactyl-Coa, And Adp
Organism: Alphaproteobacteria bacterium
Method: X-RAY DIFFRACTION Resolution:1.98 Å Release Date: 2024-10-02 Classification: LYASE Ligands: MG, 8FL, UQ3, ADP, EDO, ACY, PO4, CL, ACE, PEG |

