Search Count: 36
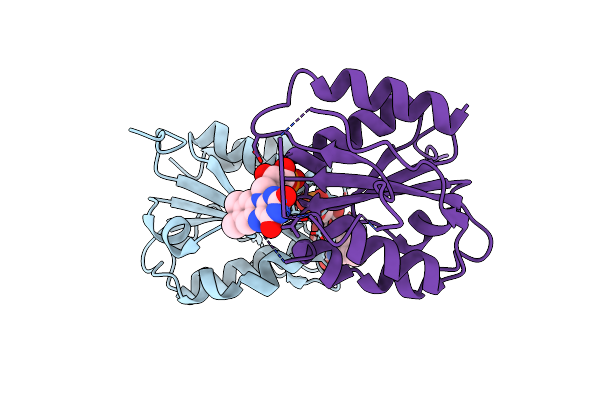 |
X-Ray Crystal Structure Of Fmn-Bound Long-Chain Flavodoxin From Rhodopseudomonas Palustris
Organism: Rhodopseudomonas palustris
Method: X-RAY DIFFRACTION Release Date: 2024-03-13 Classification: FLAVOPROTEIN Ligands: FMN |
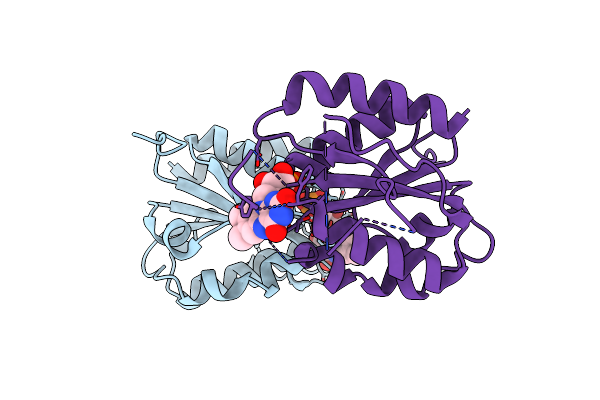 |
Room Temperature X-Ray Crystal Structure Of Fmn-Bound Long-Chain Flavodoxin From Rhodopseudomonas Palustris
Organism: Rhodopseudomonas palustris
Method: X-RAY DIFFRACTION Resolution:2.86 Å Release Date: 2024-03-13 Classification: FLAVOPROTEIN Ligands: FMN |
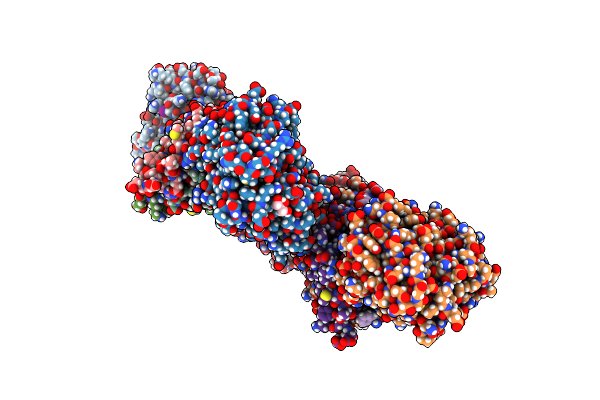 |
Organism: Bacillus wiedmannii, Bacillus phage vmy22
Method: X-RAY DIFFRACTION Resolution:2.45 Å Release Date: 2023-06-21 Classification: PROTEIN BINDING Ligands: GOL, IOD |
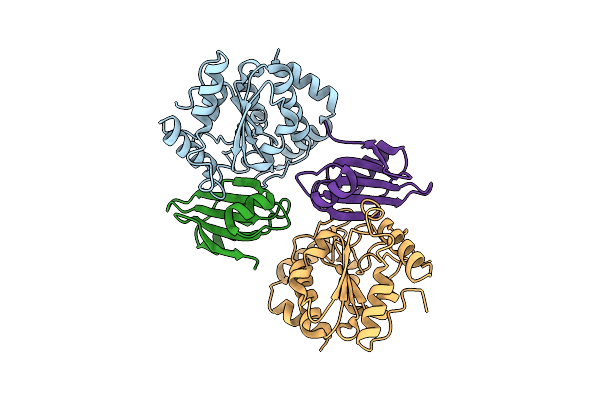 |
Organism: Bacillus phage vb_bpum-bpsp, Staphylococcus aureus
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2023-06-21 Classification: PROTEIN BINDING |
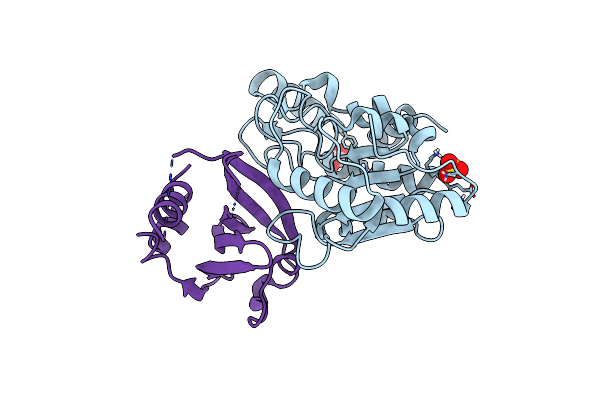 |
Organism: Staphylococcus aureus, Macrococcus caseolyticus
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2023-06-21 Classification: PROTEIN BINDING Ligands: EDO, SO4 |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.76 Å Release Date: 2022-08-10 Classification: TRANSCRIPTION Ligands: L8L |
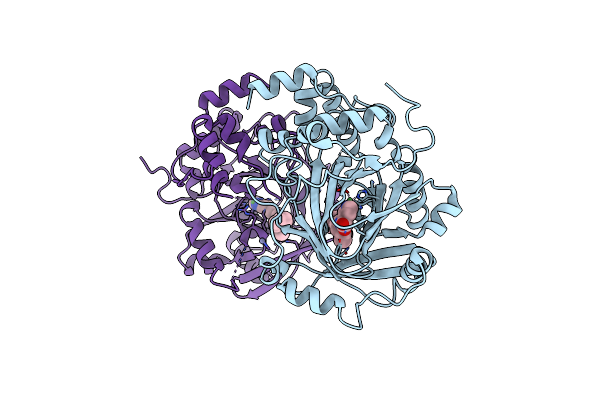 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2022-02-02 Classification: OXIDOREDUCTASE/OXIDOREDUCTASE inhibitor Ligands: NI, 9DJ, ZN |
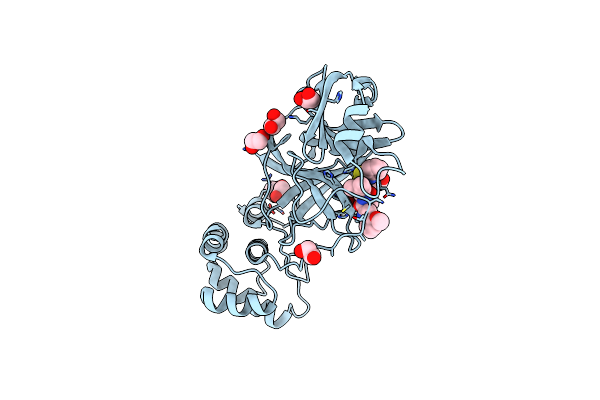 |
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2021-06-23 Classification: HYDROLASE Ligands: HUR, EDO |
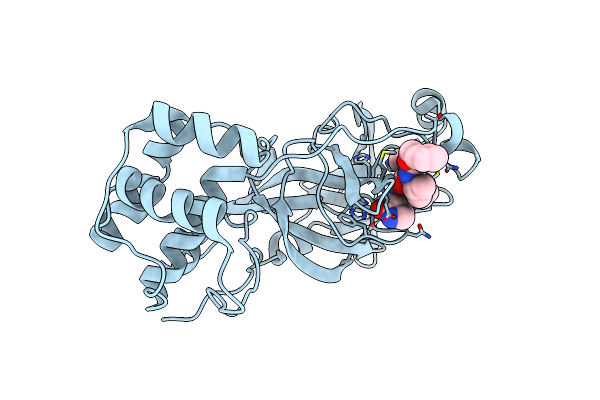 |
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2021-06-23 Classification: HYDROLASE Ligands: HUO |
 |
Organism: Cylindrotheca fusiformis
Method: SOLUTION NMR Release Date: 2016-12-21 Classification: STRUCTURAL PROTEIN |
 |
Structure Of E. Coli Beta-Glucuronidase Bound With A Novel, Potent Inhibitor 1-((6,8-Dimethyl-2-Oxo-1,2-Dihydroquinolin-3-Yl)Methyl)-1-(2-Hydroxyethyl)-3-(4-Hydroxyphenyl)Thiourea
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.39 Å Release Date: 2015-10-14 Classification: Hydrolase/Hydrolase Inhibitor Ligands: 57Z |
 |
Structure Of The Pscd4-Domain Of The Cell Wall Protein Pleuralin-1 From The Diatom Cylindrotheca Fusiformis
Organism: Cylindrotheca fusiformis
Method: SOLUTION NMR Release Date: 2015-02-25 Classification: STRUCTURAL PROTEIN |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.62 Å Release Date: 2014-12-10 Classification: HYDROLASE/HYDROLASE Inhibitor Ligands: SO4, 8H8, GOL |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2014-12-10 Classification: HYDROLASE/HYDROLASE Inhibitor Ligands: 3M8, SO4, GOL |
 |
Crystal Structure Of Dodecylphosphocholine-Solubilized Nccx From Cupriavidus Metallidurans 31A
Organism: Cupriavidus metallidurans
Method: X-RAY DIFFRACTION Resolution:3.12 Å Release Date: 2014-10-01 Classification: METAL BINDING PROTEIN Ligands: ZN, PO4, PC |
 |
Crystal Structure Of Streptococcus Agalactiae Beta-Glucuronidase In Space Group I222
Organism: Streptococcus agalactiae
Method: X-RAY DIFFRACTION Resolution:2.59 Å Release Date: 2014-09-17 Classification: HYDROLASE Ligands: MG |
 |
Crystal Structure Of Streptococcus Agalactiae Beta-Glucuronidase In Space Group P21212
Organism: Streptococcus agalactiae
Method: X-RAY DIFFRACTION Resolution:2.29 Å Release Date: 2014-09-17 Classification: HYDROLASE Ligands: MG |
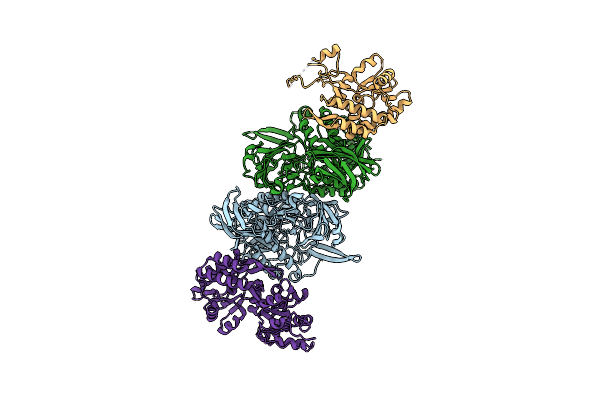 |
Organism: Clostridium perfringens, Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.26 Å Release Date: 2014-09-17 Classification: HYDROLASE |
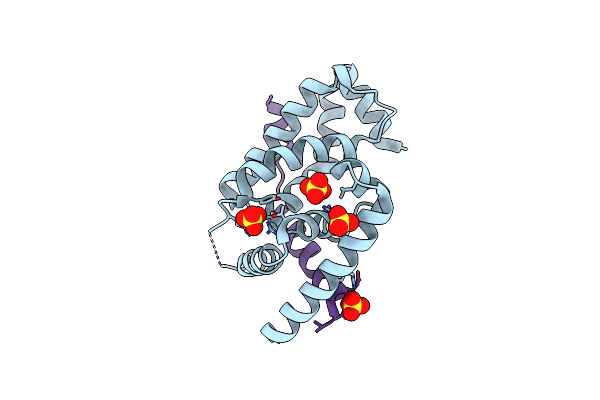 |
Organism: Cupriavidus metallidurans ch34
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2014-04-30 Classification: TRANSCRIPTION Ligands: SO4, CL |
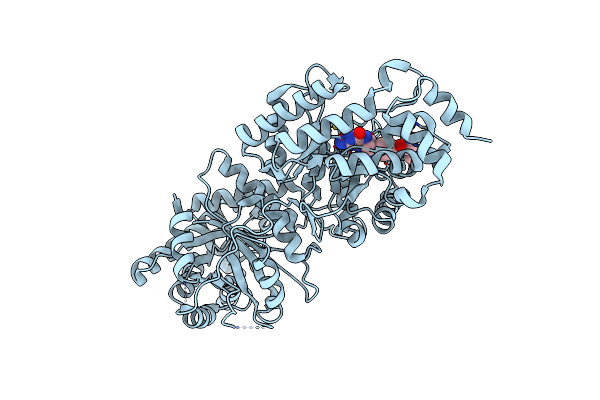 |
Crystal Structure Of Human 5-Methyltetrahydrofolate-Homocysteine Methyltransferase, The Homocysteine And Folate Binding Domains
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2013-11-27 Classification: TRANSFERASE Ligands: THG |

