Search Count: 1,300
 |
Crystal Structure Of Saccharomyces Cerevisiae Cross-Linked Sfh1 (K197C, F233C) Mutant In Complex With Phosphatidylethanolamine
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Release Date: 2025-11-05 Classification: LIPID BINDING PROTEIN Ligands: PTY, SO4, GOL |
 |
Crystal Structure Of Saccharomyces Cerevisiae Sec14P In Complex With Phosphatidylcholine
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Release Date: 2025-11-05 Classification: LIPID BINDING PROTEIN Ligands: POV, GOL, EDO, PEG |
 |
Crystal Structure Of A Ternary Complex Of Saccharomyces Cerevisiae Sec14 With A Small Molecule Inhibitor And Phosphatidylcholine
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Release Date: 2025-11-05 Classification: LIPID BINDING PROTEIN Ligands: POV, IUO |
 |
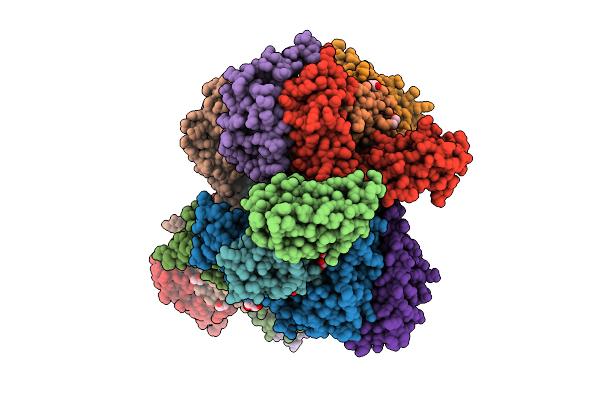
Organism: Staphylococcus aureus subsp. aureus mu50
Method: X-RAY DIFFRACTION Release Date: 2025-09-10 Classification: LIGASE Ligands: LYS, GOL |
 |
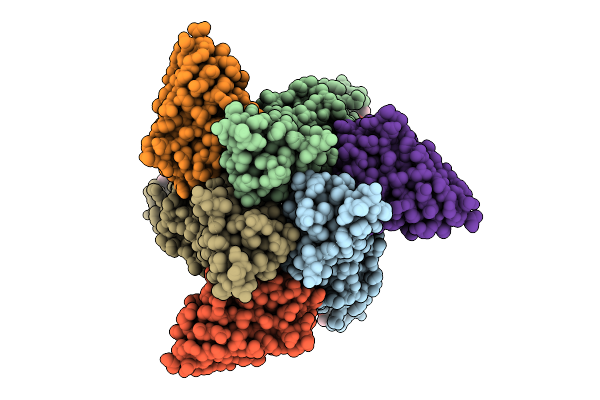
Organism: Homo sapiens, Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-07-23 Classification: IMMUNE SYSTEM Ligands: GOL |
 |
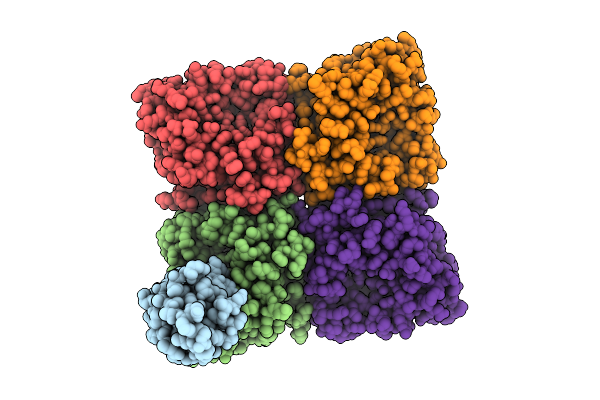
Cryo-Em Structure Of Influenza B/Washington/02/2019 Virus Hemagglutinin In Complex With Single-Domain Antibody Hvhh-69.
Organism: Influenza b virus, Lama glama
Method: ELECTRON MICROSCOPY Release Date: 2025-07-09 Classification: ANTIVIRAL PROTEIN Ligands: NAG |
 |
Cryo-Em Structure Of Influenza B/Washington/02/2019 Virus Neuraminidase In Complex With Single-Domain Antibody Hvhh-525.
Organism: Lama glama, Influenza b virus
Method: ELECTRON MICROSCOPY Release Date: 2025-07-09 Classification: ANTIVIRAL PROTEIN Ligands: NAG |
 |
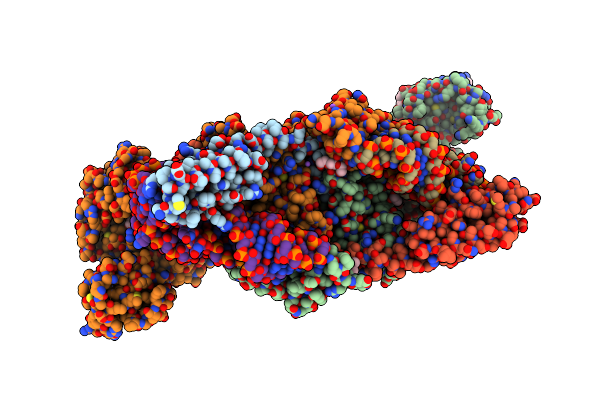
Crystal Structure Of The Complex Of M. Tuberculosis Phers With Cognate Precursor Trna And Fragment Ddd00805735
Organism: Mycobacterium tuberculosis h37rv, Mycobacterium tuberculosis (strain atcc 25618 / h37rv)
Method: X-RAY DIFFRACTION Resolution:2.51 Å Release Date: 2025-06-04 Classification: LIGASE/RNA Ligands: A1BA3, GOL, PGE, MG, NA, EPE, ACT, DMS |
 |
Crystal Structure Of The Complex Of M. Tuberculosis Phers With Cognate Precursor Trna And Fragment Ddd00107555
Organism: Mycobacterium tuberculosis h37rv
Method: X-RAY DIFFRACTION Resolution:2.05 Å Release Date: 2025-06-04 Classification: LIGASE/RNA Ligands: A1BCB, MG, ACT, NA, GOL, DMS, PGE, EPE |
 |
Crystal Structure Of The Complex Of M. Tuberculosis Phers With Cognate Precursor Trna And Fragment Ddd01008876
Organism: Mycobacterium tuberculosis h37rv
Method: X-RAY DIFFRACTION Resolution:2.45 Å Release Date: 2025-06-04 Classification: LIGASE/RNA Ligands: MG, A1BCA, ACT, NA, GOL, DMS, PGE, EPE |
 |
Mycobacterium Tuberculosis Pks13 Acyltransferase Serine Converted To Beta-Lactam Form By Cec215 Via Sufex Reaction
Organism: Mycobacterium tuberculosis h37rv
Method: X-RAY DIFFRACTION Release Date: 2025-05-21 Classification: ANTIBIOTIC Ligands: SO4, CL, PG4, EDO, DMS, GOL, PEG |
 |
Organism: Mycobacterium tuberculosis (strain atcc 25618 / h37rv)
Method: X-RAY DIFFRACTION Release Date: 2025-05-21 Classification: ANTIBIOTIC Ligands: DMS, SO4, PE5, EDO, PEG, GOL, CL, NA, P33 |
 |
Organism: Human coronavirus hku1
Method: ELECTRON MICROSCOPY Release Date: 2025-05-14 Classification: VIRAL PROTEIN Ligands: NAG |
 |
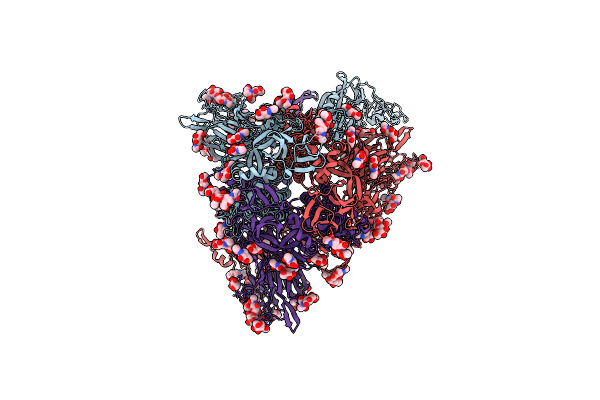
Cryo-Em Structure Of Hcov-Hku1 Glycoprotein In Complex With 9O-Acetyl Gd3 Sialoglycan (Daa State)
Organism: Human coronavirus hku1
Method: ELECTRON MICROSCOPY Release Date: 2025-05-14 Classification: VIRAL PROTEIN Ligands: NAG, A1AR1 |
 |
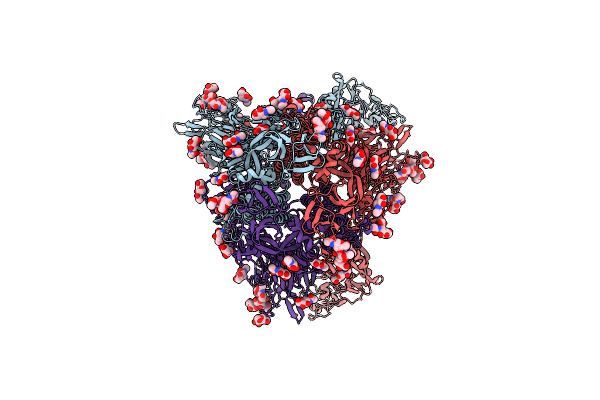
Cryo-Em Structure Of Hcov-Hku1 Glycoprotein In Complex With 9O-Acetyl Gd3 Sialoglycan (Dau State)
Organism: Human coronavirus hku1
Method: ELECTRON MICROSCOPY Release Date: 2025-05-14 Classification: VIRAL PROTEIN Ligands: NAG, A1AR1 |
 |
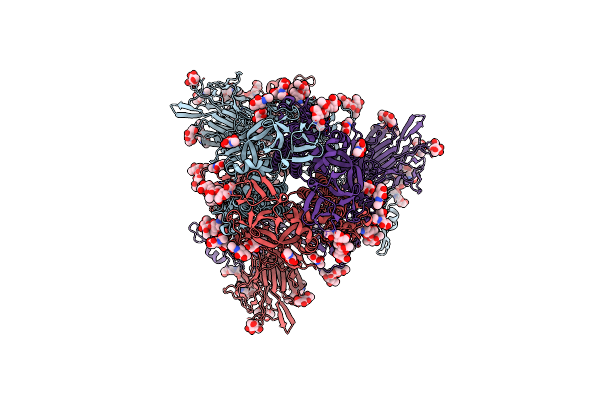
Cryo-Em Structure Of Hcov-Hku1 Glycoprotein In Complex With 9O-Acetyl Gd3 Sialoglycan (Auu State)
Organism: Human coronavirus hku1
Method: ELECTRON MICROSCOPY Release Date: 2025-05-14 Classification: VIRAL PROTEIN Ligands: NAG, A1AR2 |
 |
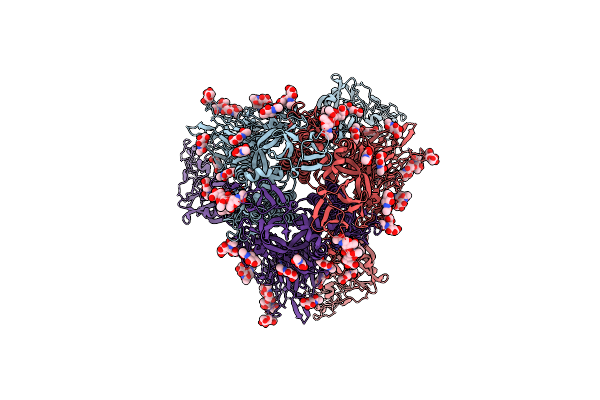
Organism: Human coronavirus hku1
Method: ELECTRON MICROSCOPY Release Date: 2025-05-14 Classification: VIRAL PROTEIN Ligands: NAG |
 |
Cryo-Em Structure Of Hcov-Hku1 Glycoprotein In Complex With 9O-Acetyl Gd3 Sialoglycan (Uuu State)
Organism: Human coronavirus hku1
Method: ELECTRON MICROSCOPY Release Date: 2025-05-14 Classification: VIRAL PROTEIN Ligands: NAG, A1AR1 |
 |
Cryo-Em Structure Of Hcov-Hku1 Glycoprotein D1 (Down State, Locally Refined)
Organism: Human coronavirus hku1
Method: ELECTRON MICROSCOPY Release Date: 2025-05-14 Classification: VIRAL PROTEIN Ligands: NAG |
 |
Organism: Human coronavirus hku1
Method: ELECTRON MICROSCOPY Release Date: 2025-05-14 Classification: VIRAL PROTEIN Ligands: NAG |

