Search Count: 337
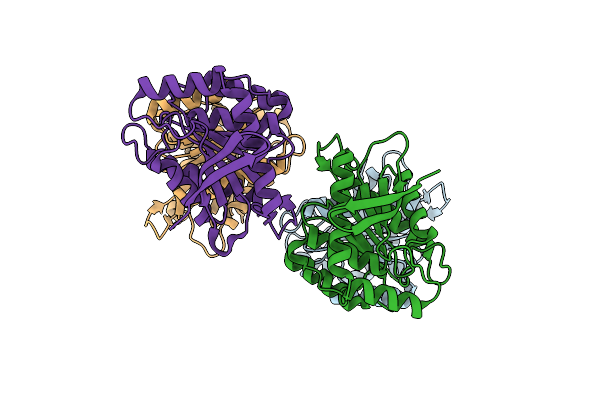 |
Organism: Hydrogenobacter thermophilus
Method: X-RAY DIFFRACTION Resolution:1.67 Å Release Date: 2025-05-14 Classification: HYDROLASE |
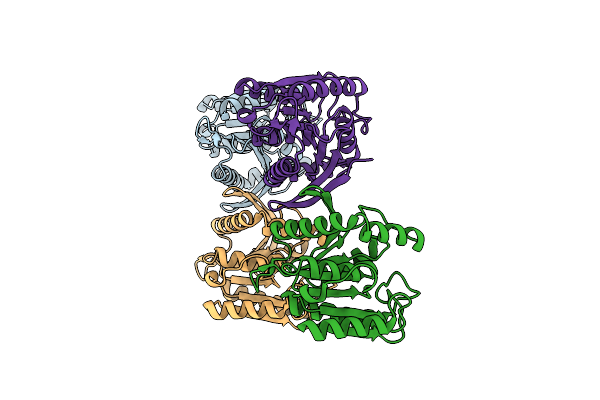 |
Crystal Structure Of Thermostable Dienelactone Hydrolase. Monoclinic Space Group.
Organism: Hydrogenobacter thermophilus
Method: X-RAY DIFFRACTION Resolution:1.79 Å Release Date: 2025-05-14 Classification: HYDROLASE |
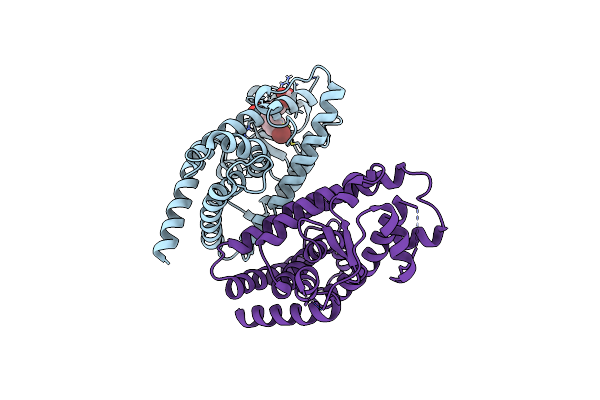 |
X-Ray Crystal Structure Of Ppar Gamma Ligand Binding Domain In Complex With Cz58
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.85 Å Release Date: 2025-05-14 Classification: TRANSCRIPTION Ligands: A1IYE |
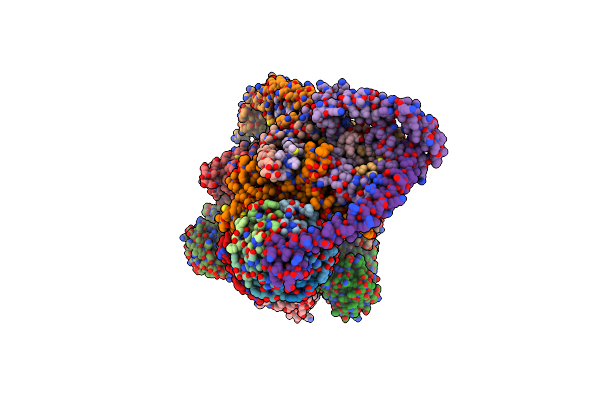 |
Organism: Artemia franciscana
Method: ELECTRON MICROSCOPY Release Date: 2025-03-26 Classification: MEMBRANE PROTEIN Ligands: ATP, MG, ADP, CDL |
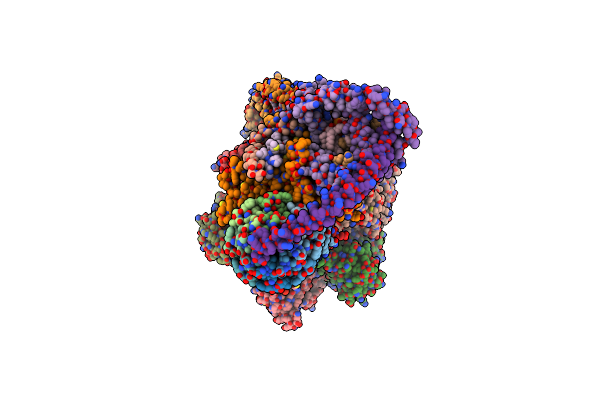 |
Organism: Artemia franciscana
Method: ELECTRON MICROSCOPY Release Date: 2025-03-26 Classification: MEMBRANE PROTEIN Ligands: ATP, MG, CDL |
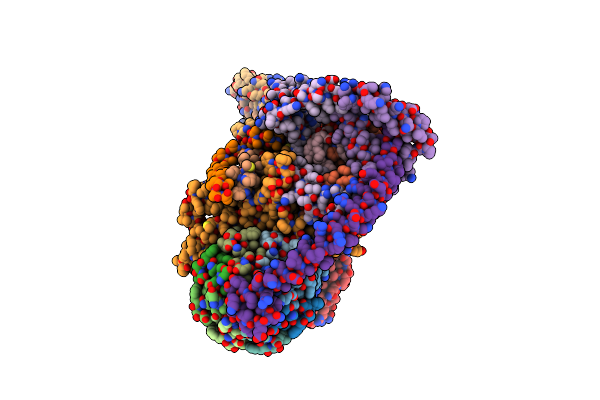 |
Organism: Artemia franciscana
Method: ELECTRON MICROSCOPY Release Date: 2025-03-26 Classification: MEMBRANE PROTEIN Ligands: CDL |
 |
N-Acetylglucosamine 6-Phosphate Dehydratase: Inhibited 6-Phosphogluconic Acid State Of Nags
Organism: Streptomyces coelicolor
Method: X-RAY DIFFRACTION Resolution:1.69 Å Release Date: 2025-02-19 Classification: SUGAR BINDING PROTEIN Ligands: SO4, 6PG |
 |
Organism: Streptomyces coelicolor
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2025-02-19 Classification: SUGAR BINDING PROTEIN Ligands: SO4 |
 |
N-Acetylglucosamine 6-Phosphate Dehydratase: Glcnac6P Substrate-Bound State Of Nags
Organism: Streptomyces coelicolor
Method: X-RAY DIFFRACTION Resolution:2.59 Å Release Date: 2025-02-19 Classification: SUGAR BINDING PROTEIN Ligands: SO4, 16G |
 |
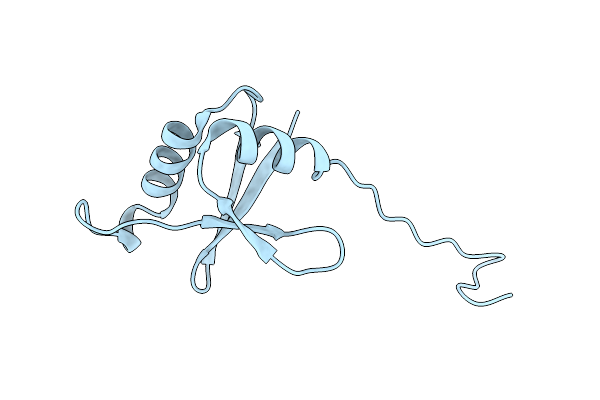
Organism: Plasmodium falciparum 3d7
Method: X-RAY DIFFRACTION Release Date: 2024-11-27 Classification: LIGASE/INHIBITOR Ligands: A1AZG, MLI, CL, NA |
 |
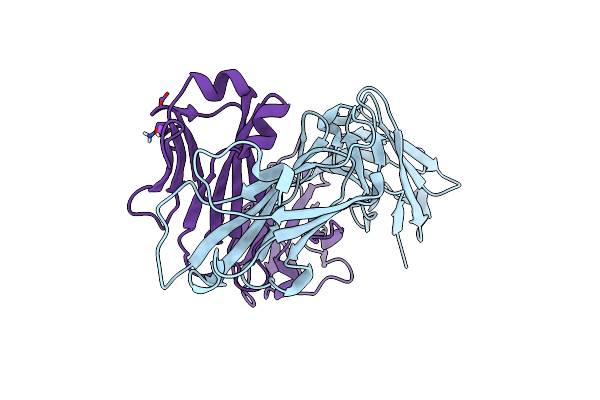
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.91 Å Release Date: 2024-11-20 Classification: PROTEIN TRANSPORT |
 |
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2024-06-26 Classification: IMMUNE SYSTEM |
 |
Organism: Escherichia coli, Escherichia phage mu
Method: ELECTRON MICROSCOPY Release Date: 2024-06-19 Classification: DNA BINDING PROTEIN Ligands: LRL, MG |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.78 Å Release Date: 2023-09-27 Classification: CELL CYCLE |
 |
Structure Of Hdac6 Zinc-Finger Ubiquitin Binding Domain In Complex With 3-(3-(2-(Methylamino)-2-Oxoethyl)-4-Oxo-3,4-Dihydroquinazolin-2-Yl)Propanoic Acid
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2023-05-03 Classification: HYDROLASE/INHIBITOR Ligands: ZN, ZU6 |
 |
Structure Of Hdac6 Zinc-Finger Ubiquitin Binding Domain In Complex With 3-(3-(2-(Benzylamino)-2-Oxoethyl)-4-Oxo-3,4-Dihydroquinazolin-2-Yl)Propanoic Acid
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2023-05-03 Classification: HYDROLASE/INHIBITOR Ligands: ZN, ZU9 |
 |
Structure Of Hdac6 Zinc-Finger Ubiquitin Binding Domain In Complex With Sgc-Ubd253 Chemical Probe
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.62 Å Release Date: 2023-05-03 Classification: HYDROLASE/INHIBITOR Ligands: ZN, ZUE |
 |
Catalytic Domain Of Udp-Glucose Glycoprotein Glucosyltransferase From Chaetomium Thermophilum In Complex With The 5-[(Morpholin-4-Yl)Methyl]Quinolin-8-Ol Inhibitor
Organism: Chaetomium thermophilum
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2023-04-26 Classification: TRANSFERASE Ligands: JM3, NAG, MAN |
 |
Crystal Structure Of Septin1 - Septin2 Heterocomplex From Drosophila Melanogaster
Organism: Drosophila melanogaster
Method: X-RAY DIFFRACTION Resolution:2.38 Å Release Date: 2023-01-11 Classification: CELL CYCLE Ligands: GDP, GTP, MG |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.65 Å Release Date: 2022-12-07 Classification: RNA BINDING PROTEIN Ligands: 9JT |

