Search Count: 691
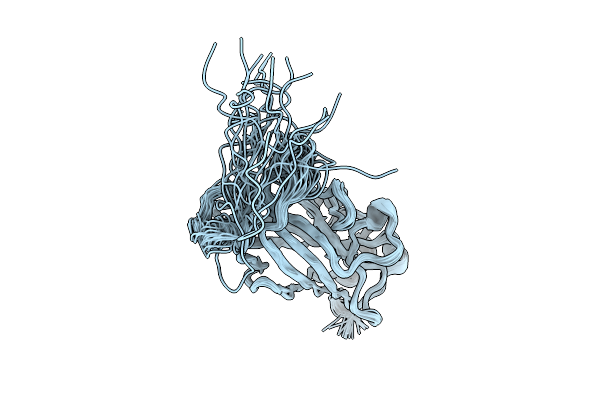 |
Structural Characterization Of The Type 3 Fimbrial Subunit Mrka From Klebsiella Pneumoniae By Solution Nmr Spectroscopy
Organism: Klebsiella pneumoniae
Method: SOLUTION NMR Release Date: 2025-11-05 Classification: CELL ADHESION |
 |
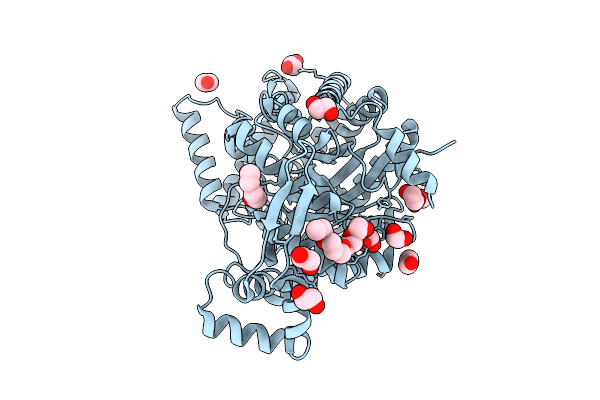
Crystal Structure Of Tagatose 4-Epimerase With One Glycerol From Thermoprotei Archaeon
Organism: Thermoprotei archaeon
Method: X-RAY DIFFRACTION Release Date: 2025-11-05 Classification: ISOMERASE Ligands: EDO, PEG, 1PG, NI, GOL |
 |
Organism: Thermoprotei archaeon
Method: X-RAY DIFFRACTION Release Date: 2025-11-05 Classification: ISOMERASE Ligands: EDO, PEG, 1PG |
 |
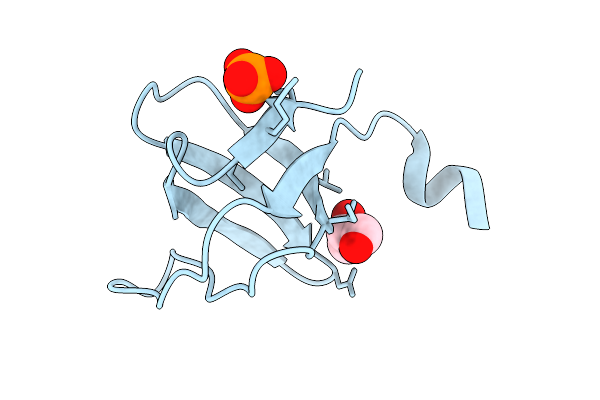
Organism: Homo sapiens, Camelidae
Method: X-RAY DIFFRACTION Release Date: 2025-10-29 Classification: IMMUNE SYSTEM Ligands: GOL |
 |
Organism: Homo sapiens, Camelidae
Method: X-RAY DIFFRACTION Release Date: 2025-10-29 Classification: IMMUNE SYSTEM |
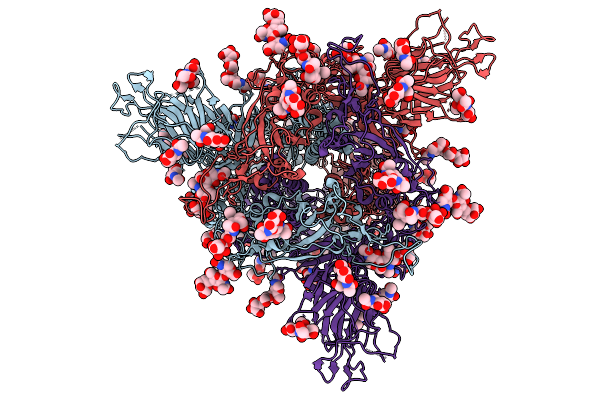 |
Organism: Sarbecovirus sp. rhgb07
Method: ELECTRON MICROSCOPY Release Date: 2025-09-24 Classification: VIRAL PROTEIN Ligands: NAG |
 |
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Release Date: 2025-08-06 Classification: SIGNALING PROTEIN Ligands: GOL, PO4 |
 |
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Release Date: 2025-08-06 Classification: SIGNALING PROTEIN Ligands: GOL, PO4 |
 |
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Release Date: 2025-08-06 Classification: SIGNALING PROTEIN Ligands: GOL, PO4 |
 |
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Release Date: 2025-08-06 Classification: SIGNALING PROTEIN |
 |
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Release Date: 2025-08-06 Classification: STRUCTURAL PROTEIN Ligands: GOL |
 |
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Release Date: 2025-08-06 Classification: STRUCTURAL PROTEIN Ligands: SO4 |
 |
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Release Date: 2025-08-06 Classification: STRUCTURAL PROTEIN Ligands: SO4 |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-05-21 Classification: LYASE Ligands: EDO, ACT, ZN, A1IBT |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-05-21 Classification: LYASE Ligands: EDO, ZN, JKE |
 |
Organism: Microbacterium testaceum
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2025-04-23 Classification: LYASE Ligands: RB |
 |
Microbacterium Testaceum C-Glucosyl Deglycosidase (Cgd), Wild Type (Cryo-Em)
Organism: Microbacterium testaceum
Method: ELECTRON MICROSCOPY Release Date: 2025-04-23 Classification: LYASE Ligands: MN |
 |
Organism: Neisseria gonorrhoeae fa 1090
Method: X-RAY DIFFRACTION Resolution:3.50 Å Release Date: 2025-04-16 Classification: DNA BINDING PROTEIN |
 |
Crystal Structure Of The Complex Of Human Carbonic Anhydrase I With 4-Sulfamoylphenyl 3-(P-Tolylthio)Propanoate
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-03-26 Classification: LYASE Ligands: ACT, ZN, A1H6C |
 |

