Search Count: 18
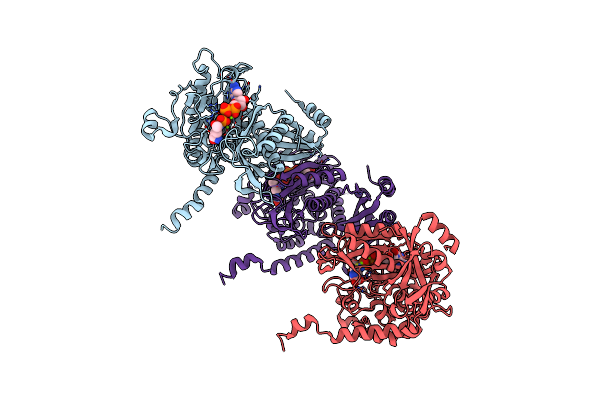 |
Structure Of Paenibacillus Polymyxa Gs Bound To Met-Sox-P-Adp (Transition State Complex) To 1.98 Angstom
Organism: Paenibacillus polymyxa
Method: X-RAY DIFFRACTION Resolution:1.98 Å Release Date: 2022-06-29 Classification: LIGASE Ligands: ADP, P3S, MG |
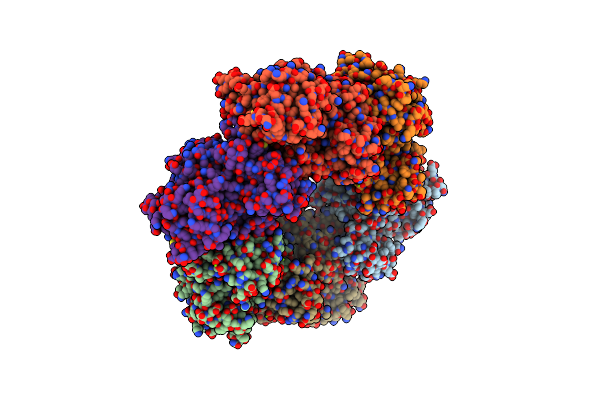 |
Crystal Structure Of S. Aureus Glutamine Synthetase In Met-Sox-P/Adp Transition State Complex
Organism: Staphylococcus aureus
Method: X-RAY DIFFRACTION Resolution:2.92 Å Release Date: 2022-06-29 Classification: LIGASE Ligands: ADP, P3S, MG, SO4 |
 |
Organism: Staphylococcus aureus
Method: X-RAY DIFFRACTION Resolution:2.35 Å Release Date: 2022-06-29 Classification: DNA BINDING PROTEIN/DNA Ligands: CA |
 |
Organism: Listeria monocytogenes, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:3.45 Å Release Date: 2022-06-29 Classification: DNA BINDING PROTEIN/DNA |
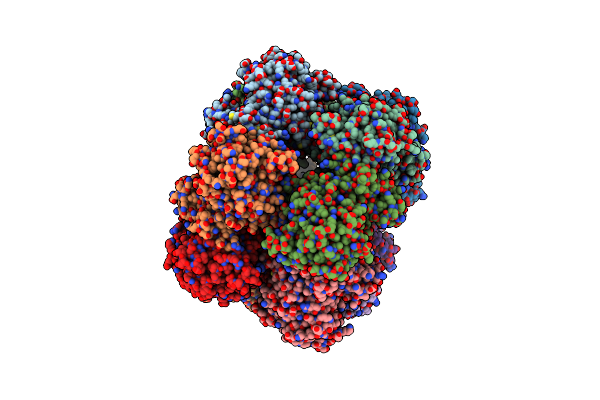 |
Crystal Structure Of The Listeria Monocytogenes Gs-Met-Sox-P- Adp Complex To 3.5 Angstrom
Organism: Listeria monocytogenes
Method: X-RAY DIFFRACTION Resolution:3.50 Å Release Date: 2022-06-29 Classification: LIGASE/INHIBITOR Ligands: ADP, P3S |
 |
Organism: Staphylococcus aureus
Method: ELECTRON MICROSCOPY Release Date: 2022-06-29 Classification: BIOSYNTHETIC PROTEIN, LIGASE Ligands: MG, GLN |
 |
Organism: Staphylococcus aureus
Method: ELECTRON MICROSCOPY Release Date: 2022-06-29 Classification: BIOSYNTHETIC PROTEIN, LIGASE |
 |
Organism: Listeria monocytogenes
Method: ELECTRON MICROSCOPY Release Date: 2022-06-29 Classification: BIOSYNTHETIC PROTEIN, LIGASE Ligands: MG, GLN |
 |
Organism: Paenibacillus polymyxa
Method: ELECTRON MICROSCOPY Release Date: 2022-06-29 Classification: BIOSYNTHETIC PROTEIN, LIGASE Ligands: MG, GLN |
 |
Organism: Paenibacillus polymyxa
Method: ELECTRON MICROSCOPY Release Date: 2022-06-29 Classification: BIOSYNTHETIC PROTEIN, LIGASE Ligands: MG, GLN |
 |
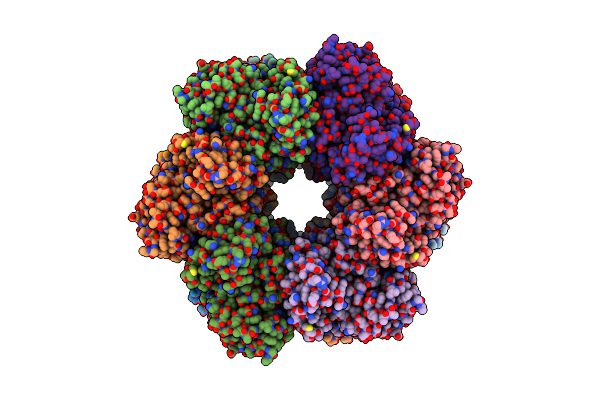
Organism: Bacillus subtilis
Method: ELECTRON MICROSCOPY Release Date: 2022-06-29 Classification: BIOSYNTHETIC PROTEIN, LIGASE Ligands: MG, GLN |
 |
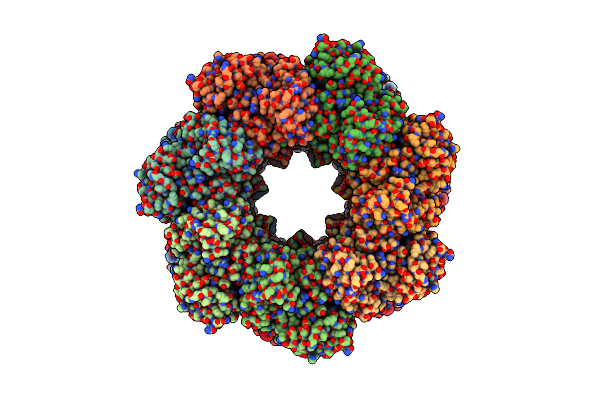
Organism: Paenibacillus polymyxa
Method: ELECTRON MICROSCOPY Release Date: 2022-06-29 Classification: BIOSYNTHETIC PROTEIN Ligands: MG |
 |
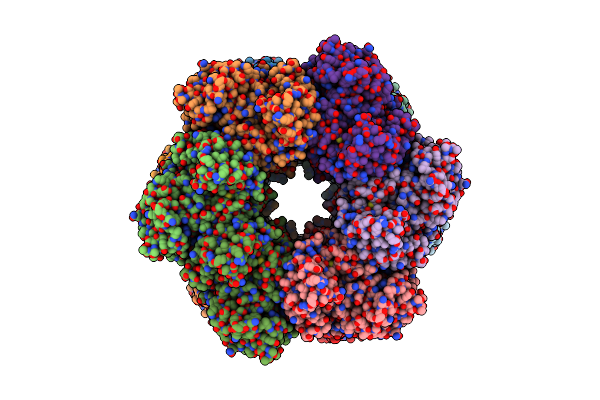
Organism: Listeria monocytogenes
Method: ELECTRON MICROSCOPY Release Date: 2022-06-29 Classification: BIOSYNTHETIC PROTEIN Ligands: MG |
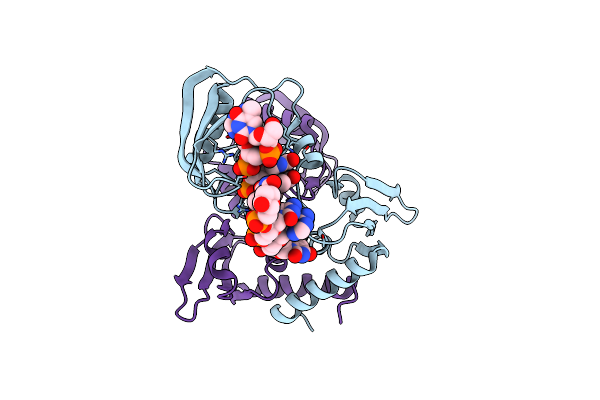 |
Organism: Caulobacter vibrioides, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2022-06-01 Classification: TRANSCRIPTION |
 |
Organism: Caulobacter vibrioides (strain atcc 19089 / cb15), Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.48 Å Release Date: 2022-06-01 Classification: TRANSCRIPTION Ligands: SO4 |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.83 Å Release Date: 2011-10-19 Classification: ISOMERASE |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2011-01-12 Classification: OXIDOREDUCTASE Ligands: SO4 |
 |
Alternative Conformations Of The X Region Of Human Protein Disulphide-Isomerase Modulate Exposure Of The Substrate Binding B' Domain
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2008-09-30 Classification: ISOMERASE Ligands: SO4 |

