Search Count: 21
 |
Organism: Azotobacter vinelandii dj
Method: X-RAY DIFFRACTION Release Date: 2025-12-03 Classification: METAL BINDING PROTEIN Ligands: SF4, S5Q, PEG, EDO, 1PE, PGE, MG, PG4 |
 |
Organism: Geobacter metallireducens
Method: ELECTRON MICROSCOPY Release Date: 2025-11-05 Classification: METAL BINDING PROTEIN Ligands: SF4, S5Q |
 |
Organism: Geobacter metallireducens
Method: ELECTRON MICROSCOPY Release Date: 2025-11-05 Classification: METAL BINDING PROTEIN Ligands: SF4, S5Q |
 |
Crystal Structure Of The Complex Formed Between The Radical Sam Protein Chlb And The Leader Region Of Its Precursor Substrate Chla
Organism: Fischerella
Method: X-RAY DIFFRACTION Release Date: 2025-06-25 Classification: OXIDOREDUCTASE Ligands: SF4, FMT, SAH, TRS, ZN |
 |
Crystal Structure Of The Complex Formed Between The Radical Sam Protein Chlb And The R3A Mutant Of Chla
Organism: Fischerella
Method: X-RAY DIFFRACTION Release Date: 2025-06-25 Classification: OXIDOREDUCTASE Ligands: SF4, SAH, ACT |
 |
Crystal Structure Of Ethylene Glycol-Bound Glycerol Dehydrogenase From Klebsiella Pneumoniae
Organism: Klebsiella pneumoniae
Method: X-RAY DIFFRACTION Resolution:2.09 Å Release Date: 2024-05-29 Classification: OXIDOREDUCTASE Ligands: ZN, EDO, GOL |
 |
Crystal Structure Of Ethylene Glycol-Bound Glycerol Dehydrogenase From Klebsiella Pneumoniae
Organism: Klebsiella pneumoniae
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2024-05-29 Classification: OXIDOREDUCTASE Ligands: ZN, NAD, EDO, TLA |
 |
Crystal Structure Of The P97-N/D1 Hexamer In Complex With Six P47-Ubx Domains
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2023-12-20 Classification: PROTEIN BINDING Ligands: ADP |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2023-07-12 Classification: PROTEIN BINDING Ligands: SAM |
 |
Crystal Structure Of P97 N/D1 In Complex With A Valosin-Containing Protein Methyltransferase
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2023-07-12 Classification: PROTEIN BINDING Ligands: SAH, GOL, FMT, ACT, ADP, EPE |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.99 Å Release Date: 2022-09-21 Classification: PROTEIN BINDING Ligands: ZN |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.72 Å Release Date: 2022-09-21 Classification: PROTEIN BINDING |
 |
Organism: Acinetobacter baumannii
Method: X-RAY DIFFRACTION Resolution:2.65 Å Release Date: 2022-05-18 Classification: HYDROLASE/HYDROLASE INHIBITOR Ligands: FDX |
 |
Organism: Acinetobacter baumannii
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2022-05-18 Classification: HYDROLASE/HYDROLASE INHIBITOR Ligands: FDX |
 |
Organism: Acinetobacter baumannii
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2022-05-18 Classification: HYDROLASE/HYDROLASE INHIBITOR Ligands: FDX |
 |
Organism: Acinetobacter baumannii
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2022-05-04 Classification: HYDROLASE Ligands: IM2 |
 |
Solution Structure Of An Intramolecular G-Quadruplex Containing A Duplex Bulge
|
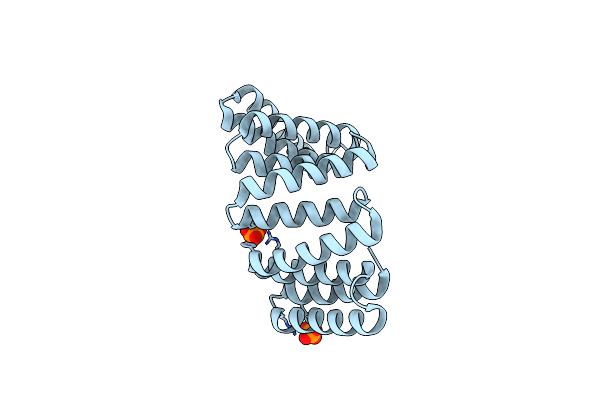 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.25 Å Release Date: 2017-10-11 Classification: MOTOR PROTEIN Ligands: PO4 |
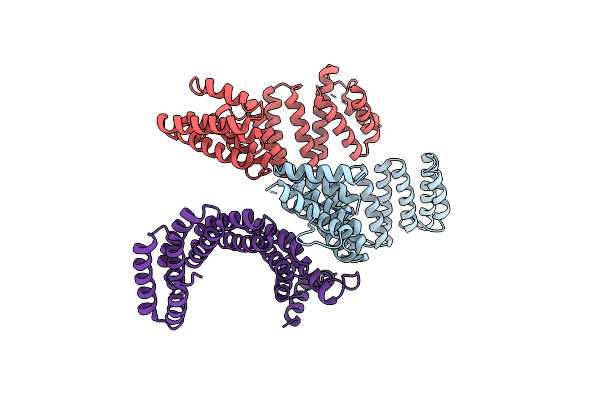 |
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:3.40 Å Release Date: 2017-10-11 Classification: MOTOR PROTEIN |
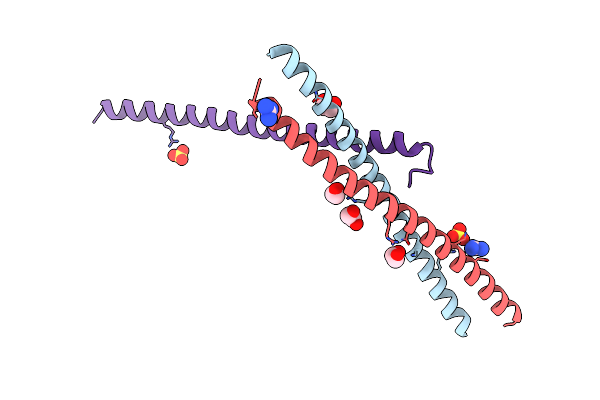 |
Crystallographic Structure Of The Lzii Fragment (Anti-Parallel Orientation) From Jip3
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.06 Å Release Date: 2015-06-10 Classification: PROTEIN BINDING Ligands: SO4, EDO, GAI |

