Search Count: 95
 |
 |
 |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-07-16 Classification: RNA BINDING PROTEIN |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-07-16 Classification: RNA BINDING PROTEIN |
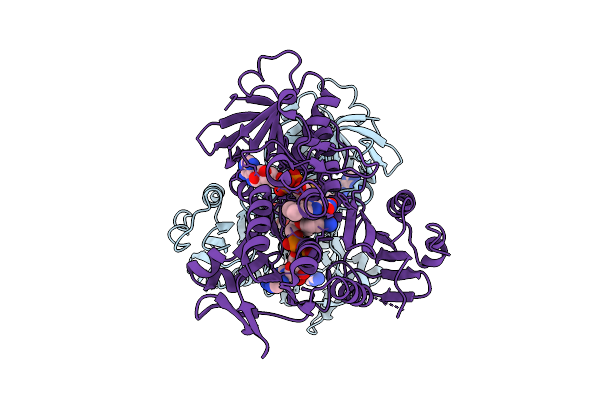 |
Interaction With Ak2A Links Aifm1 To Cellular Energy Metabolism. The Cryo-Em Structure Of Dimeric Aifm1 Without Any Binding Partner.
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-07-09 Classification: FLAVOPROTEIN Ligands: FAD, NAD |
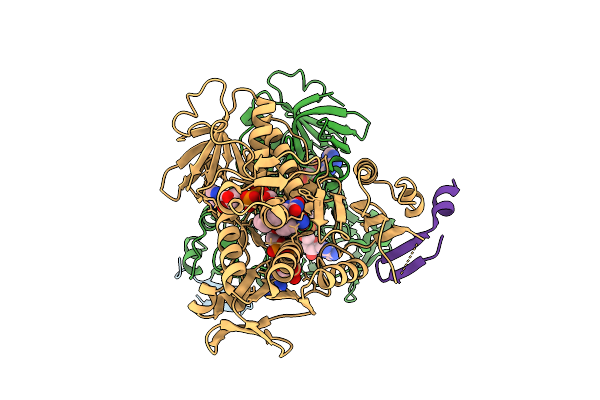 |
Interaction With Ak2A Links Aifm1 To Cellular Energy Metabolism. The Cryo-Em Structure Of Dimeric Aifm1 Engaged To Mia40.
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-07-09 Classification: FLAVOPROTEIN Ligands: FAD, NAD |
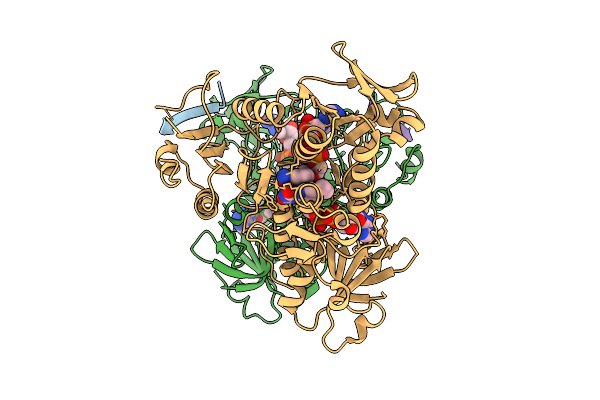 |
Interaction With Ak2A Links Aifm1 To Cellular Energy Metabolism. The Cryo-Em Structure Of Dimeric Aifm1 Bound By Ak2A.
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-07-09 Classification: FLAVOPROTEIN Ligands: FAD, NAD |
 |
Organism: Escherichia coli (strain k12), Saccharomyces cerevisiae (strain atcc 204508 / s288c), Homo sapiens
Method: ELECTRON MICROSCOPY Resolution:2.30 Å Release Date: 2025-03-12 Classification: RNA BINDING PROTEIN Ligands: MG, ZN, D3T |
 |
The Human Line-1 Orf2P Target-Primed Reverse Transcription Complex With The Fingers Domain In A Closed Conformation
Organism: Escherichia coli (strain k12), Saccharomyces cerevisiae (strain atcc 204508 / s288c), Homo sapiens
Method: ELECTRON MICROSCOPY Resolution:2.50 Å Release Date: 2025-03-12 Classification: RNA BINDING PROTEIN Ligands: MG, ZN, D3T |
 |
The Human Line-1 Orf2P Target-Primed Reverse Transcription Complex With The Fingers Domain In An Open Conformation
Organism: Escherichia coli (strain k12), Saccharomyces cerevisiae (strain atcc 204508 / s288c), Homo sapiens
Method: ELECTRON MICROSCOPY Resolution:2.45 Å Release Date: 2025-03-12 Classification: RNA BINDING PROTEIN Ligands: ZN |
 |
Human Line-1 Orf2P Target-Primed Reverse Transcription Complex With En Domain Resolved
Organism: Escherichia coli (strain k12), Saccharomyces cerevisiae (strain atcc 204508 / s288c), Homo sapiens
Method: ELECTRON MICROSCOPY Resolution:3.10 Å Release Date: 2025-03-12 Classification: RNA BINDING PROTEIN Ligands: MG, ZN, D3T |
 |
Organism: Arabidopsis thaliana
Method: ELECTRON MICROSCOPY Resolution:3.00 Å Release Date: 2025-02-26 Classification: MEMBRANE PROTEIN Ligands: K |
 |
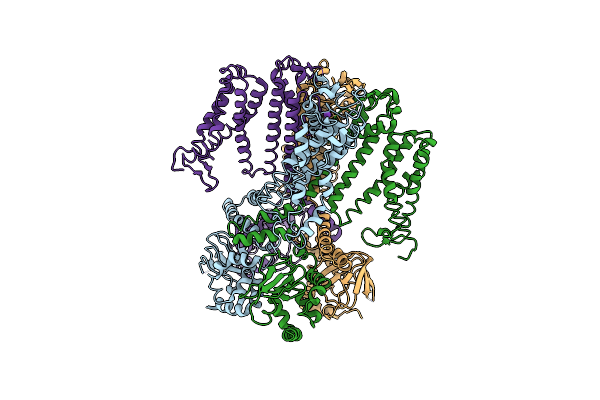
Organism: Arabidopsis thaliana
Method: ELECTRON MICROSCOPY Release Date: 2025-02-26 Classification: MEMBRANE PROTEIN Ligands: K |
 |
Organism: Arabidopsis thaliana
Method: ELECTRON MICROSCOPY Resolution:2.60 Å Release Date: 2025-02-26 Classification: MEMBRANE PROTEIN Ligands: K |
 |
Organism: Arabidopsis thaliana
Method: ELECTRON MICROSCOPY Release Date: 2025-02-26 Classification: MEMBRANE PROTEIN Ligands: K |
 |
Organism: Homo sapiens, Influenza a virus
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2024-12-18 Classification: IMMUNE SYSTEM Ligands: PO4 |
 |
Organism: Homo sapiens, Influenza a virus
Method: X-RAY DIFFRACTION Resolution:2.65 Å Release Date: 2024-12-18 Classification: IMMUNE SYSTEM |
 |
Organism: Homo sapiens, Influenza a virus
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2024-12-18 Classification: IMMUNE SYSTEM Ligands: EDO, NA, PO4 |
 |
Crystal Structure Of A Hla-B*35:01-Np6 Epitope From 1977 H1N1 Influenza Strain
Organism: Homo sapiens, Influenza a virus (strain a/1977/h1n1)
Method: X-RAY DIFFRACTION Release Date: 2024-03-27 Classification: IMMUNE SYSTEM |

