Search Count: 33
 |
Crystal Structure Of Larp-Dm15 From Drosophila Melanogaster Bound To M7Gpppc
Organism: Drosophila melanogaster
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2024-01-17 Classification: RNA BINDING PROTEIN Ligands: 91P, MG |
 |
Organism: Danio rerio
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2024-01-17 Classification: RNA BINDING PROTEIN Ligands: 91P, MY6 |
 |
Organism: Thermoascus aurantiacus
Method: X-RAY DIFFRACTION Resolution:1.99 Å Release Date: 2020-07-22 Classification: HYDROLASE Ligands: ACT, SO4 |
 |
Organism: Thermoascus aurantiacus
Method: X-RAY DIFFRACTION Resolution:1.62 Å Release Date: 2020-07-22 Classification: HYDROLASE Ligands: ACT, SO4 |
 |
Organism: Thermoascus aurantiacus
Method: X-RAY DIFFRACTION Resolution:1.35 Å Release Date: 2020-07-22 Classification: HYDROLASE Ligands: SO4 |
 |
Organism: Thermoascus aurantiacus
Method: X-RAY DIFFRACTION Resolution:1.47 Å Release Date: 2020-07-22 Classification: HYDROLASE Ligands: SO4 |
 |
Organism: Thermoascus aurantiacus
Method: X-RAY DIFFRACTION Resolution:1.73 Å Release Date: 2020-07-22 Classification: HYDROLASE Ligands: ACT |
 |
Organism: Thermoascus aurantiacus
Method: X-RAY DIFFRACTION Resolution:1.47 Å Release Date: 2020-07-22 Classification: HYDROLASE Ligands: ACT, SO4 |
 |
Crystal Structure Of Kemp Eliminase Hg3 With Bound Transition State Analogue, 277K
Organism: Thermoascus aurantiacus
Method: X-RAY DIFFRACTION Resolution:1.86 Å Release Date: 2020-07-22 Classification: HYDROLASE Ligands: 6NT, SO4 |
 |
Crystal Structure Of Kemp Eliminase Hg3.3B With Bound Transition State Analogue, 277K
Organism: Thermoascus aurantiacus
Method: X-RAY DIFFRACTION Resolution:1.42 Å Release Date: 2020-07-22 Classification: HYDROLASE Ligands: 6NT, SO4 |
 |
Crystal Structure Of Kemp Eliminase Hg3.7 With Bound Transition State Analogue, 277K
Organism: Thermoascus aurantiacus
Method: X-RAY DIFFRACTION Resolution:1.39 Å Release Date: 2020-07-22 Classification: HYDROLASE Ligands: 6NT, SO4 |
 |
Crystal Structure Of Kemp Eliminase Hg3.14 With Bound Transition State Analogue, 277K
Organism: Thermoascus aurantiacus
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2020-07-22 Classification: HYDROLASE Ligands: 6NT, SO4 |
 |
Crystal Structure Of Kemp Eliminase Hg3.17 With Bound Transition State Analog, 277K
Organism: Thermoascus aurantiacus
Method: X-RAY DIFFRACTION Resolution:1.77 Å Release Date: 2020-07-22 Classification: HYDROLASE Ligands: 6NT |
 |
Crystal Structure Of Kemp Eliminase Hg4 With Bound Transition State Analogue, 277K
Organism: Thermoascus aurantiacus
Method: X-RAY DIFFRACTION Resolution:1.46 Å Release Date: 2020-07-22 Classification: HYDROLASE Ligands: 6NT, SO4 |
 |
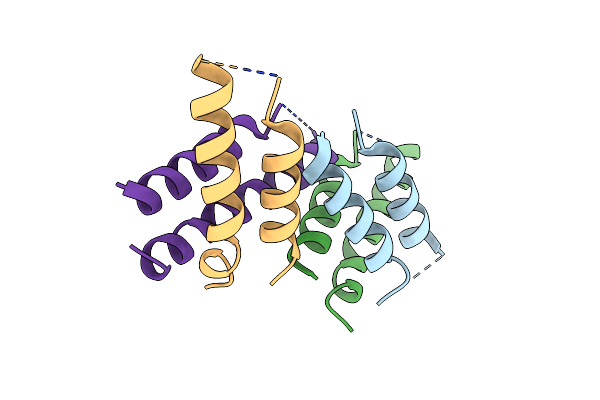
Crystal Structure Of Human Transferrin Receptor In Complex With A Cystine-Dense Peptide
Organism: Homo sapiens, Monosiga brevicollis
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2020-04-15 Classification: TRANSPORT PROTEIN/SIGNALING PROTEIN Ligands: MES, GOL, NAG |
 |
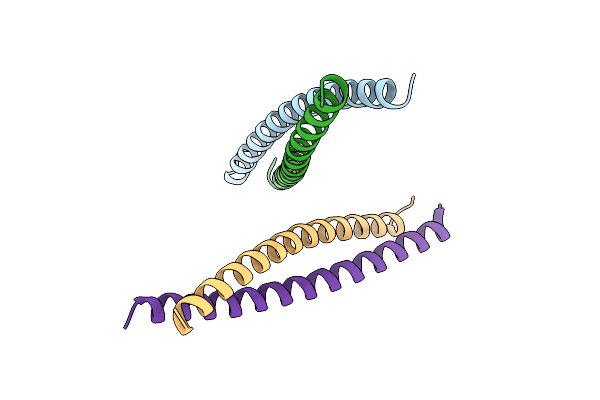
Crystal Structure Of An Apo Transferrin-Receptor-Binding Cystine-Dense Peptide
Organism: Monosiga brevicollis
Method: X-RAY DIFFRACTION Resolution:2.55 Å Release Date: 2020-04-15 Classification: SIGNALING PROTEIN |
 |
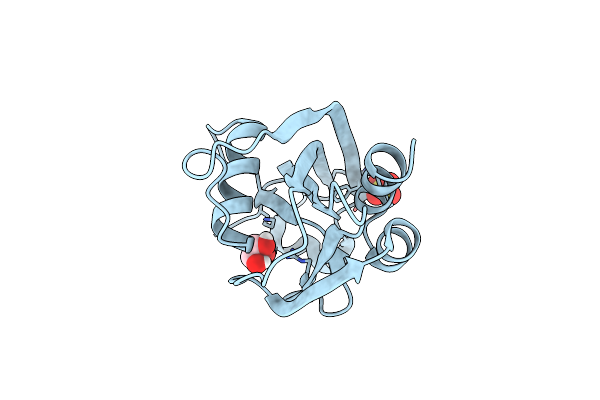
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:1.92 Å Release Date: 2017-11-08 Classification: SIGNALING PROTEIN |
 |
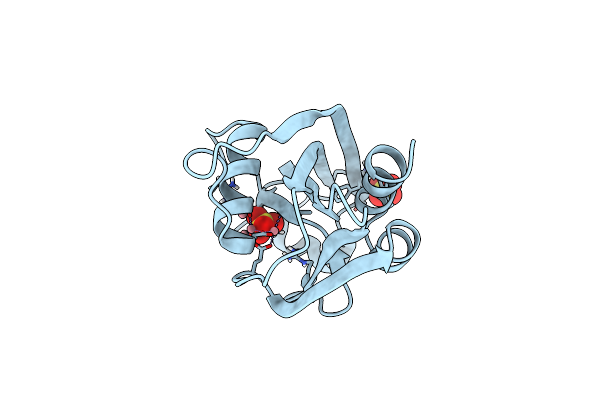
Organism: Escherichia coli o157:h7
Method: X-RAY DIFFRACTION Resolution:1.94 Å Release Date: 2017-06-21 Classification: HYDROLASE Ligands: SO4, GOL |
 |
Organism: Escherichia coli o157:h7
Method: X-RAY DIFFRACTION Resolution:1.79 Å Release Date: 2017-06-21 Classification: HYDROLASE Ligands: SO4, CO |
 |
Organism: Escherichia coli o157:h7
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2017-06-21 Classification: HYDROLASE Ligands: SO4, NI |

