Search Count: 180
 |
Crystal Structure Of Ice Binding Protein From Candidatus Cryosericum Odellii Smc5
Organism: Candidatus cryosericum odellii
Method: X-RAY DIFFRACTION Resolution:1.45 Å Release Date: 2025-09-03 Classification: RECOMBINATION |
 |
Crystal Structure Of Ice Binding Protein From Candidatus Cryosericum Odellii Smc5
Organism: Candidatus cryosericum odellii
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2025-09-03 Classification: RECOMBINATION |
 |
Crystal Structure Of An Ice-Binding Protein From Candidatus Cryosericum Odellii
Organism: Candidatus cryosericum odellii
Method: X-RAY DIFFRACTION Resolution:1.83 Å Release Date: 2025-09-03 Classification: PROTEIN BINDING |
 |
Crystal Structure Of Engineered Ice-Binding Protein (M74I And A97V) From Candidatus Cryosericum Odellii Strain Smc5_169
Organism: Candidatus cryosericum odellii
Method: X-RAY DIFFRACTION Resolution:1.45 Å Release Date: 2025-09-03 Classification: PROTEIN BINDING |
 |
Crystal Structure Of Transcriptional Regulator (Nrpr) From Streptococcus Salivarius K12
Organism: Streptococcus salivarius k12
Method: X-RAY DIFFRACTION Resolution:2.59 Å Release Date: 2025-05-21 Classification: TRANSCRIPTION |
 |
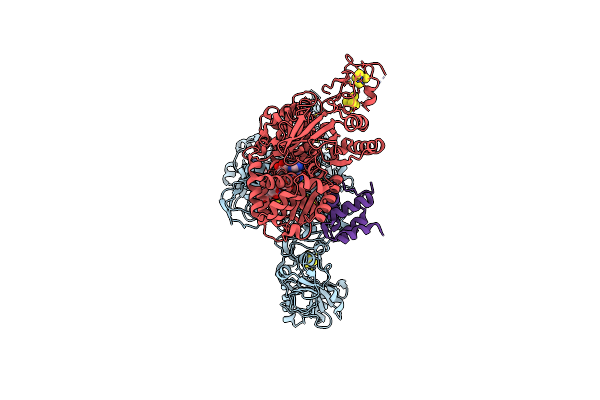
Organism: Thermococcus sibiricus
Method: ELECTRON MICROSCOPY Resolution:2.85 Å Release Date: 2025-05-07 Classification: ELECTRON TRANSPORT Ligands: SF4, FAD, FES, FMN, ZN |
 |
Organism: Thermococcus sibiricus
Method: ELECTRON MICROSCOPY Resolution:2.76 Å Release Date: 2025-05-07 Classification: ELECTRON TRANSPORT Ligands: SF4, FAD, FES, FMN, NAD, ZN |
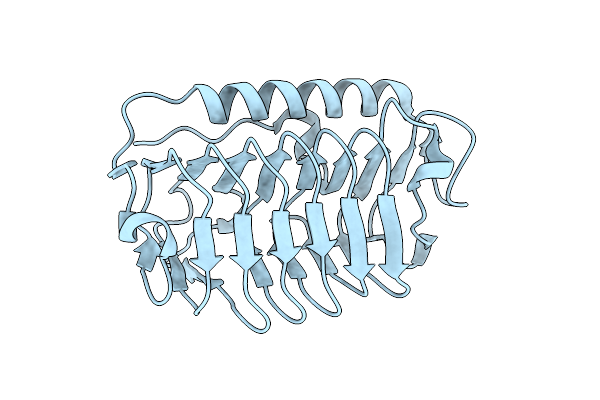 |
Organism: Flavobacterium frigoris ps1
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2024-11-13 Classification: ANTIFREEZE PROTEIN |
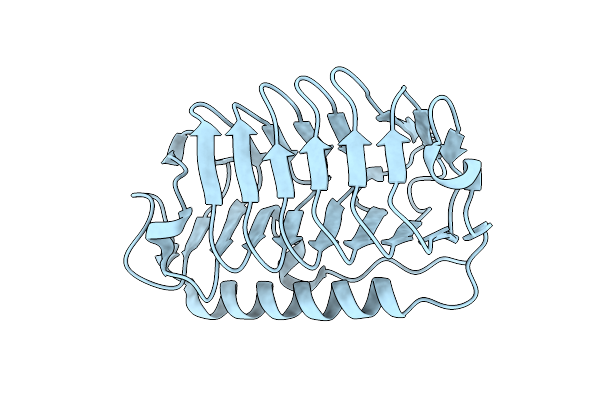 |
Organism: Flavobacterium frigoris ps1
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2024-11-13 Classification: ANTIFREEZE PROTEIN |
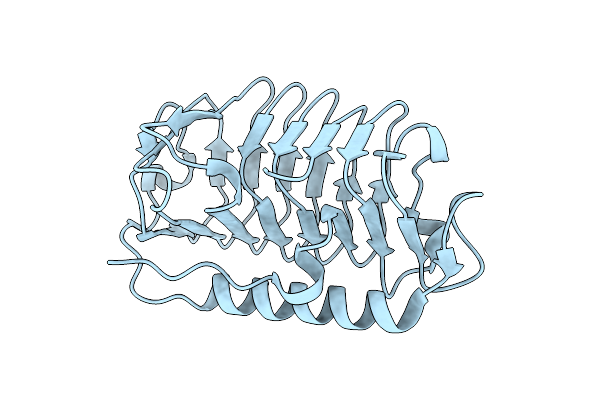 |
Organism: Flavobacterium frigoris ps1
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2024-11-13 Classification: ANTIFREEZE PROTEIN |
 |
Organism: Flavobacterium frigoris ps1
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2024-11-13 Classification: ANTIFREEZE PROTEIN |
 |
Organism: Microcystis aeruginosa nies-88
Method: X-RAY DIFFRACTION Resolution:1.94 Å Release Date: 2024-06-19 Classification: TRANSFERASE Ligands: PIS, MG, BTB |
 |
Solution Conformations Of A 12-Mer Peptide Bearing A Natural N-Hydrophobic Triangle
Organism: Synthetic construct
Method: SOLUTION NMR Release Date: 2024-06-05 Classification: PROTEIN BINDING |
 |
Solution Structure Of A 12-Mer Peptide Bearing A Bicyclic Asx Motif Mimic (Bamm) As A Synthetic N-Cap
Organism: Synthetic construct
Method: SOLUTION NMR Release Date: 2024-06-05 Classification: PEPTIDE BINDING PROTEIN Ligands: ZBR |
 |
Crystal Structure Of Sulfur Transferase From Frondihabitans Sp. Pamc28461 Crystallized In The P1 Space Group
Organism: Frondihabitans sp.
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2024-04-24 Classification: TRANSFERASE |
 |
Crystal Structure Of Sulfur Transferase From Frondihabitans Sp. Pamc28461 Crystallized In The I21 Space Group
Organism: Frondihabitans sp.
Method: X-RAY DIFFRACTION Resolution:2.38 Å Release Date: 2024-04-24 Classification: TRANSFERASE |
 |
Crystal Structure Of Hiv-1 Lm/Ht Clade A/E Crf01 Gp120 Core In Complex With Tfh-Ii-128
Organism: Human immunodeficiency virus 1
Method: X-RAY DIFFRACTION Resolution:2.42 Å Release Date: 2024-03-20 Classification: VIRAL PROTEIN/INHIBITOR Ligands: NAG, Z6F, EPE |
 |
Crystal Structure Of Hiv-1 Lm/Ht Clade A/E Crf01 Gp120 Core In Complex With Tfh-Ii-151
Organism: Human immunodeficiency virus 1
Method: X-RAY DIFFRACTION Resolution:2.65 Å Release Date: 2024-03-20 Classification: VIRAL PROTEIN/INHIBITOR Ligands: NAG, Z7N, EPE, CL |
 |
Organism: Synthetic construct, Lama glama, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-01-03 Classification: MEMBRANE PROTEIN Ligands: YUX |
 |
Cryo-Em Structure Of T/F100 Sosip.664 Hiv-1 Env Trimer With Lmhs Mutations In Complex With 8Anc195 And 10-1074
Organism: Human immunodeficiency virus 1, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2023-11-08 Classification: VIRAL PROTEIN/IMMUNE SYSTEM Ligands: NAG |

