Search Count: 9
 |
Organism: Proteus mirabilis
Method: X-RAY DIFFRACTION Resolution:1.62 Å Release Date: 2022-07-06 Classification: SUGAR BINDING PROTEIN Ligands: CL |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2022-07-06 Classification: SUGAR BINDING PROTEIN Ligands: IOD |
 |
Organism: Proteus mirabilis
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2022-07-06 Classification: SUGAR BINDING PROTEIN Ligands: FUL, CL |
 |
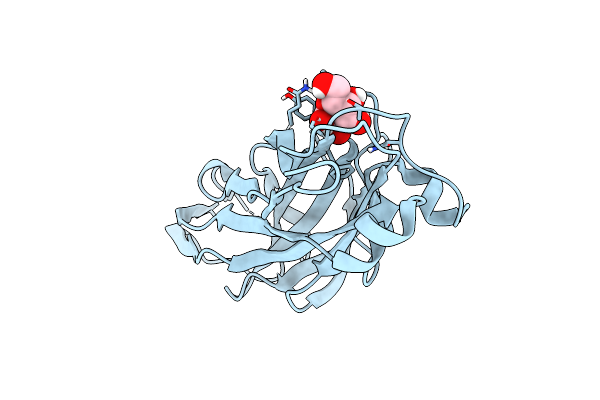
Crystal Structure Of The Ucad Lectin-Binding Domain In Complex With Glucose
Organism: Proteus mirabilis
Method: X-RAY DIFFRACTION Resolution:1.78 Å Release Date: 2022-07-06 Classification: SUGAR BINDING PROTEIN Ligands: BGC, CL |
 |
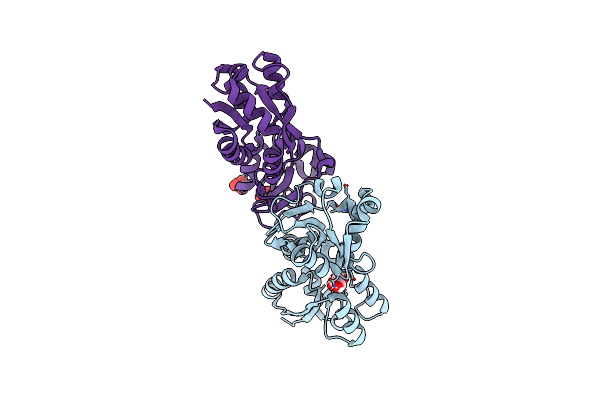
Crystal Structure Of The Ucad Lectin-Binding Domain In Complex With Galactose
Organism: Proteus mirabilis
Method: X-RAY DIFFRACTION Resolution:1.72 Å Release Date: 2022-07-06 Classification: SUGAR BINDING PROTEIN Ligands: GLA, CL |
 |
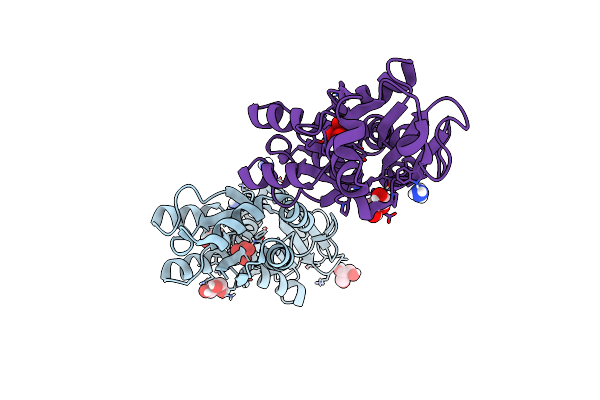
Crystal Structure Of The Molybdate-Binding Periplasmic Protein Moda From The Bacteria Pseudomonsa Aeruginosa In Ligand-Free Form
Organism: Pseudomonas aeruginosa pa1
Method: X-RAY DIFFRACTION Resolution:1.78 Å Release Date: 2022-05-04 Classification: METAL BINDING PROTEIN Ligands: GOL, NH4, SO4 |
 |
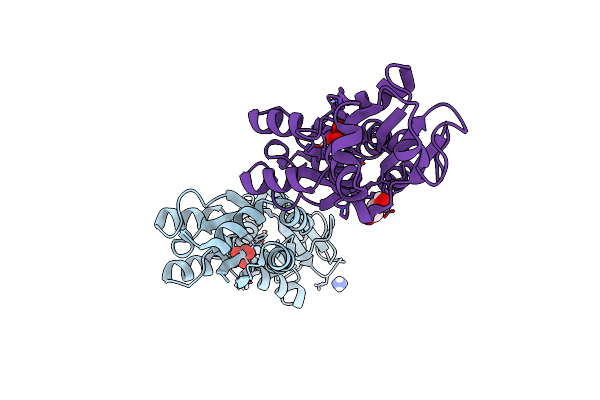
Crystal Structure Of The Molybdate-Binding Periplasmic Protein Moda From The Bacteria Pseudomonsa Aeruginosa In Chromate-Bound Form
Organism: Pseudomonas aeruginosa pa1
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2022-05-04 Classification: METAL BINDING PROTEIN Ligands: CQ4, NH4, GOL |
 |
Crystal Structure Of The Molybdate-Binding Periplasmic Protein Moda From The Bacteria Pseudomonsa Aeruginosa In Molybdate-Bound Form
Organism: Pseudomonas aeruginosa pa1
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2022-05-04 Classification: METAL BINDING PROTEIN Ligands: NH4, MOO, GOL |
 |
Crystal Structure Of The Molybdate-Binding Periplasmic Protein Moda From The Bacteria Pseudomonsa Aeruginosa In Tungstate-Bound Form
Organism: Pseudomonas aeruginosa pa1
Method: X-RAY DIFFRACTION Resolution:2.16 Å Release Date: 2022-05-04 Classification: METAL BINDING PROTEIN Ligands: WO4, NH4 |

