Search Count: 36
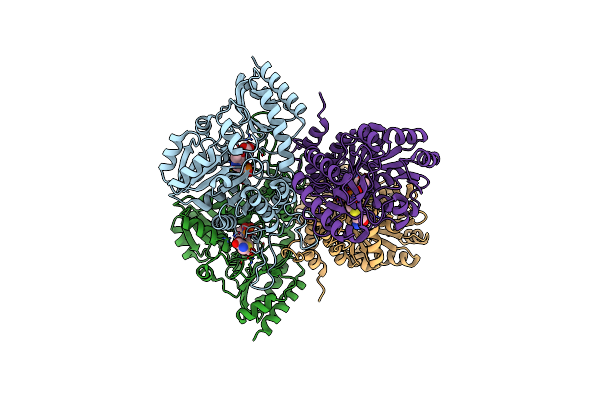 |
Crystal Structure Of Cystathionine Gamma Synthase From Xanthomonas Oryzae Pv. Oryzae In Complex With Homolanthionine
Organism: Xanthomonas oryzae pv. oryzae kacc 10331
Method: X-RAY DIFFRACTION Resolution:2.39 Å Release Date: 2020-12-09 Classification: BIOSYNTHETIC PROTEIN Ligands: EBO |
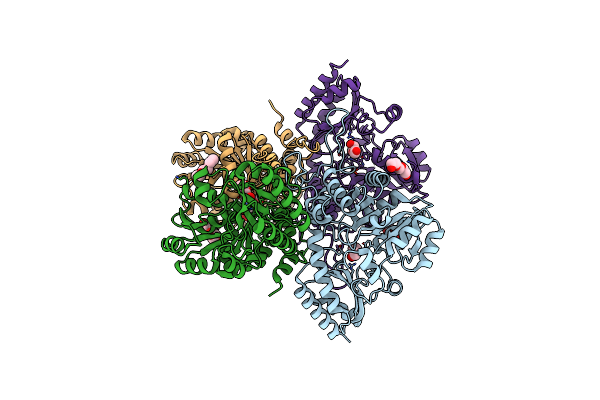 |
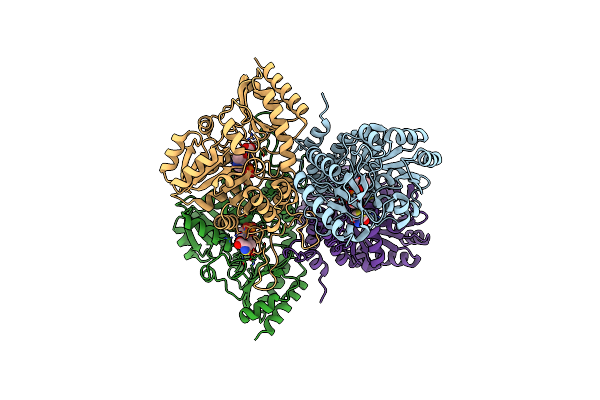
Native Structure Of Cystathionine Gamma Synthase (Xometb) From Xanthomonas Oryzae Pv. Oryzae
Organism: Xanthomonas oryzae pv. oryzae kacc 10331
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2020-11-25 Classification: BIOSYNTHETIC PROTEIN Ligands: PEG, PGE |
 |
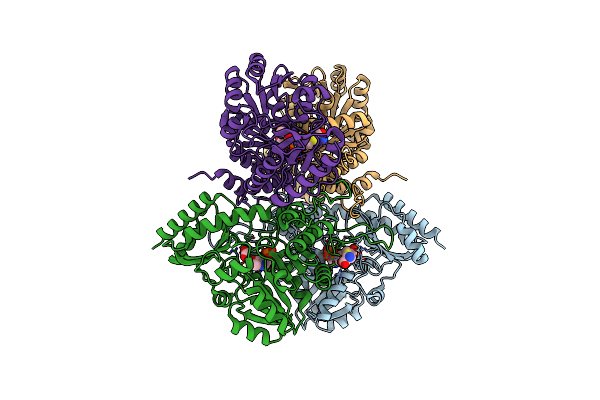
Crystal Structure Of Cystathionine Gamma Synthase From Xanthomonas Oryzae Pv. Oryzae In Complex With Aminoacrylate And Cysteine
Organism: Xanthomonas oryzae pv. oryzae (strain kacc10331 / kxo85)
Method: X-RAY DIFFRACTION Resolution:2.31 Å Release Date: 2020-11-25 Classification: BIOSYNTHETIC PROTEIN Ligands: 0JO, CYS |
 |
Crystal Structure Of Cystathionine Gamma Synthase From Xanthomonas Oryzae Pv. Oryzae In Complex With Cystathionine
Organism: Xanthomonas oryzae pv. oryzae kacc 10331
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2020-11-25 Classification: BIOSYNTHETIC PROTEIN Ligands: E9U |
 |
Organism: Bacillus cereus
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2017-09-06 Classification: OXIDOREDUCTASE |
 |
Organism: Bacillus cereus
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2017-09-06 Classification: OXIDOREDUCTASE Ligands: NAD, NA |
 |
Organism: Bacillus cereus
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2017-09-06 Classification: OXIDOREDUCTASE |
 |
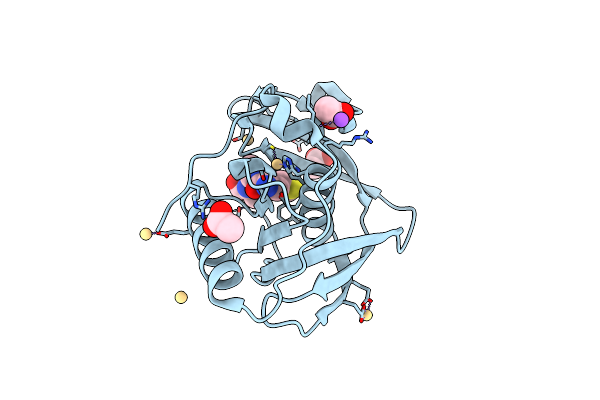
Native Structure Of Xoo1075, A Peptide Deformylase From Xanthomonas Oryzae Pv. Oryzae
Organism: Xanthomonas oryzae pv. oryzae kacc10331
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2016-10-12 Classification: HYDROLASE Ligands: CD |
 |
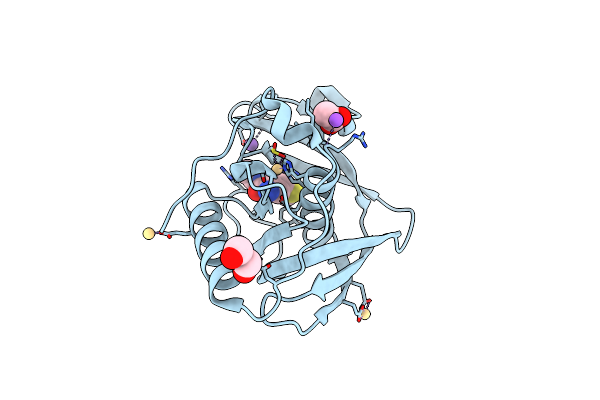
Mas Complex Structure Of Peptide Deformylase From Xanthomonas Oryzae Pv Oryzae
Organism: Xanthomonas oryzae pv. oryzae kacc10331, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2016-08-03 Classification: HYDROLASE/SUBSTRATE Ligands: CD, ACT, NA |
 |
Methionine-Alanine Complex Structure Of Peptide Deformylase From Xanthomonas Oryzae Pv. Oryzae
Organism: Xanthomonas oryzae pv. oryzae kacc10331
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2016-08-03 Classification: HYDROLASE Ligands: CD, ACT, NA, GOL, MET, ALA |
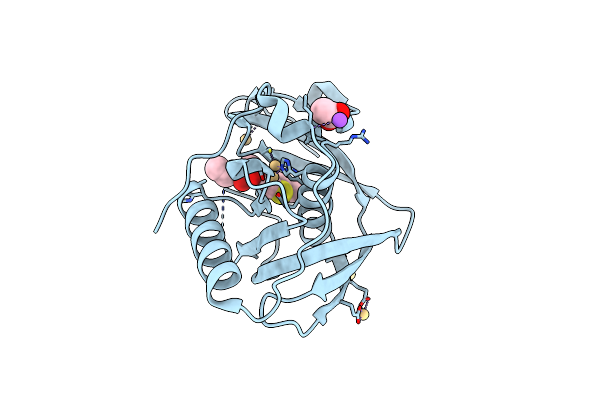 |
Structure Of Xoo1075, A Peptide Deformyase From Xanthomonas Oryzae Pv. Oryze, In Complex With Fragment 493
Organism: Xanthomonas oryzae pv. oryzae kacc10331
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2016-08-03 Classification: HYDROLASE/HYDROLASE INHIBITOR Ligands: CD, 56E, ACT, NA |
 |
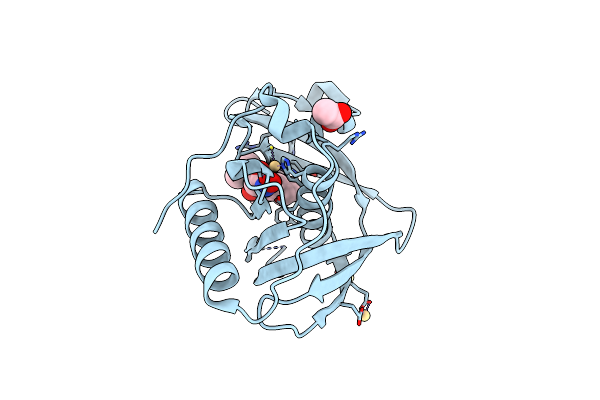
Structure Of Xoo1075, A Peptide Deformylase From Xanthomonas Oryzae Pv Oryze, In Complex With Fragment 571
Organism: Xanthomonas oryzae pv. oryzae kacc10331
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2016-08-03 Classification: HYDROLASE/HYDROLASE INHIBITOR Ligands: CD, ACT, NA, 56F |
 |
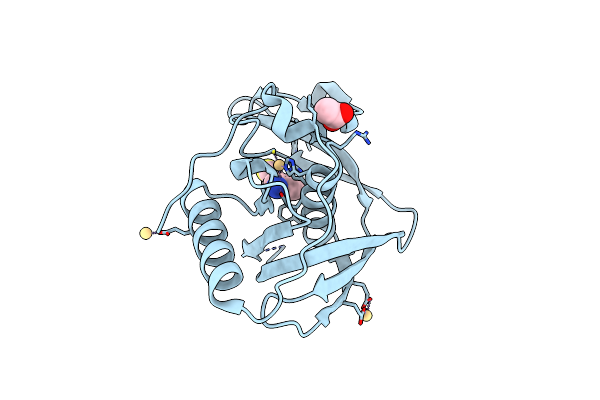
Structure Of Xoo1075, A Peptide Deformylase From Xanthomonas Oryzae Pv Oryzae, In Complex With Actinonin
Organism: Xanthomonas oryzae pv. oryzae kacc10331
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2016-08-03 Classification: HYDROLASE/HYDROLASE INHIBITOR Ligands: CD, ACT, BB2 |
 |
Structure Of Xoo1075, A Peptide Deformylase From Xanthomonas Oryzae Pv Oryzae, In Complex With Fragment 134
Organism: Xanthomonas oryzae pv. oryzae kacc10331
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2016-08-03 Classification: HYDROLASE Ligands: CD, ACT, 56K |
 |
Structure Of Xoo1075, A Peptide Deformylase From Xanthomonas Oryze Pv Oryzae, In Complex With Fragment 83
Organism: Xanthomonas oryzae pv. oryzae kacc10331
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2016-08-03 Classification: HYDROLASE Ligands: CD, ACT, 4WL, NA |
 |
Structure Of Xoo1075, A Peptide Deformylase From Xanthomonas Oryzae Pv. Oryzae, In Complex With Fragment 322
Organism: Xanthomonas oryzae pv. oryzae kacc10331
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2016-08-03 Classification: HYDROLASE Ligands: CD, ACT, 56L |
 |
Structure Of Xoo1075, A Peptide Deformylase From Xanthomonas Oryzae Pv Oryzae, In Complex With Fragment 124
Organism: Xanthomonas oryzae pv. oryzae kacc10331
Method: X-RAY DIFFRACTION Resolution:2.38 Å Release Date: 2016-08-03 Classification: HYDROLASE/HYDROLASE INHIBITOR Ligands: ACT, CD, 56T, NA |
 |
Structure Of Xoo1075, A Peptide Deformylase From Xanthomonas Oryze Pv Oryze, In Complex With Fragment 275
Organism: Xanthomonas oryzae pv. oryzae kacc10331
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2016-08-03 Classification: HYDROLASE/HYDROLASE INHIBITOR Ligands: CD, ACT, 56U, NA |
 |
Structure Of Xoo1075, A Peptide Deformylase From Xanthomonas Oryzae Pv Oryze, In Complex With Fragment 244
Organism: Xanthomonas oryzae pv. oryzae kacc10331
Method: X-RAY DIFFRACTION Resolution:2.38 Å Release Date: 2016-08-03 Classification: HYDROLASE/HYDROLASE INHIBITOR Ligands: 56V, CD, ACT, NA |
 |
Crystal Structure Of Adp Complexed D-Alanine-D-Alanine Ligase(Ddl) From Yersinia Pestis
Organism: Yersinia pestis
Method: X-RAY DIFFRACTION Resolution:2.28 Å Release Date: 2016-03-02 Classification: LIGASE |

