Search Count: 204
 |
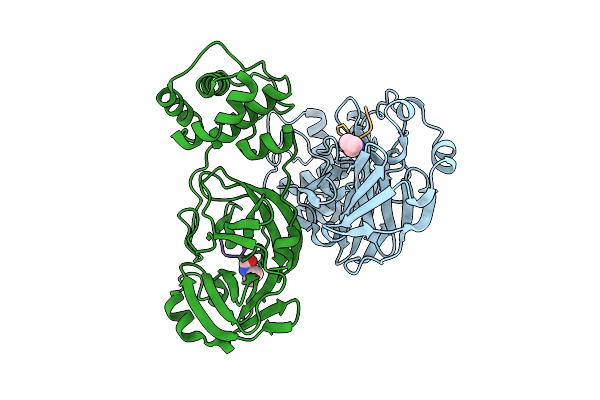
Crystal Structure Of The Moae-Like Domain Within Rv3323C From Mycobacterium Tuberculosis
Organism: Mycobacterium tuberculosis (strain atcc 25618 / h37rv)
Method: X-RAY DIFFRACTION Release Date: 2025-08-27 Classification: TRANSFERASE Ligands: MES, GOL |
 |
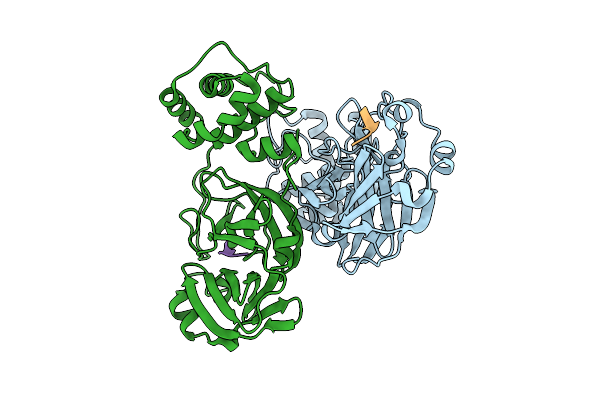
Discovery Of An Orally Bioavailable Reversible Covalent Sars-Cov-2 Mpro Inhibitor With Pan-Coronavirus Activity
Organism: Severe acute respiratory syndrome coronavirus 2, Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-08-20 Classification: HYDROLASE/INHIBOTOR Ligands: A1L7M |
 |
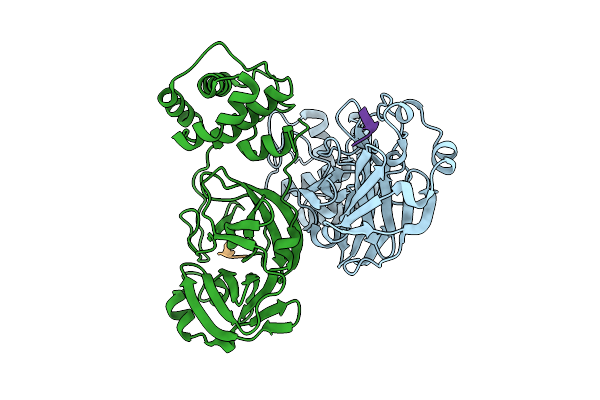
Discovery Of An Orally Bioavailable Reversible Covalent Sars-Cov-2 Mpro Inhibitor With Pan-Coronavirus Activity
Organism: Severe acute respiratory syndrome coronavirus 2, Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-08-20 Classification: HYDROLASE/INHIBITOR |
 |
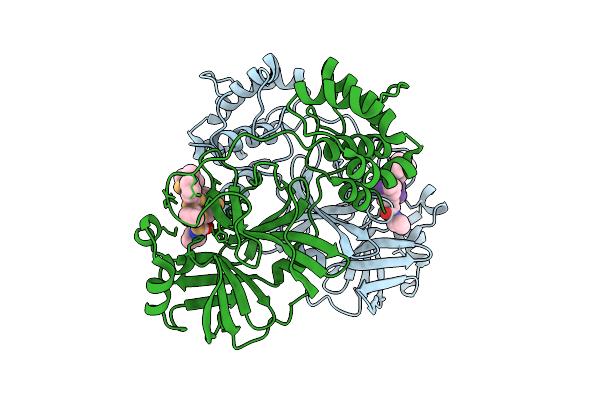
Discovery Of An Orally Bioavailable Reversible Covalent Sars-Cov-2 Mpro Inhibitor With Pan-Coronavirus Activity
Organism: Severe acute respiratory syndrome coronavirus 2, Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-08-20 Classification: HYDROLASE/INHIBITOR |
 |
Discovery Of An Orally Bioavailable Reversible Covalent Sars-Cov-2 Mpro Inhibitor With Pan-Coronavirus Activity
Organism: Severe acute respiratory syndrome coronavirus 2, Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-08-20 Classification: HYDROLASE/INHIBITOR Ligands: A1L7M |
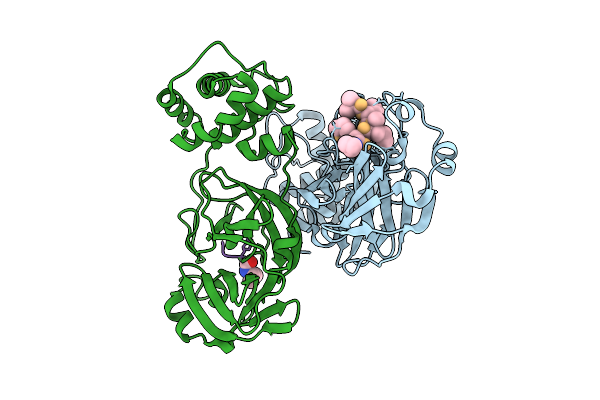 |
Discovery Of An Orally Bioavailable Reversible Covalent Sars-Cov-2 Mpro Inhibitor With Pan-Coronavirus Activity
Organism: Severe acute respiratory syndrome coronavirus 2, Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-08-20 Classification: HYDROLASE/INHIBITOR Ligands: A1L7M |
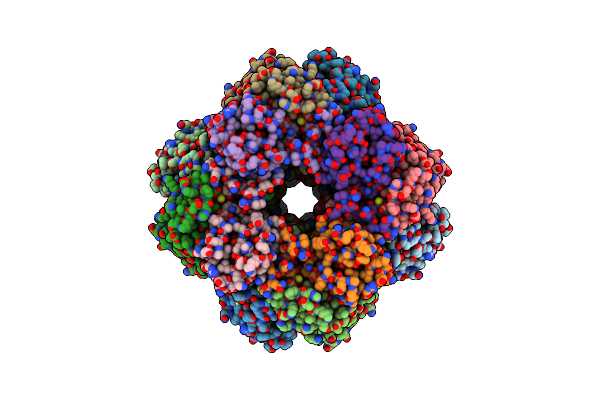 |
Cryo-Em Structure Of The Rubisco From Thermophilic Purple Bacterial Rubisco
Organism: Thermochromatium tepidum atcc 43061
Method: ELECTRON MICROSCOPY Release Date: 2025-03-05 Classification: PLANT PROTEIN |
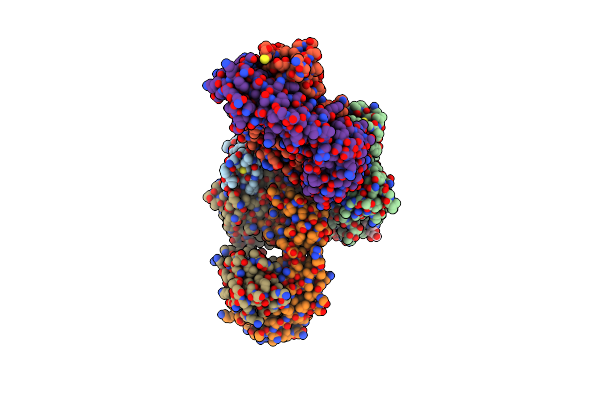 |
Organism: Severe acute respiratory syndrome coronavirus 2, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.45 Å Release Date: 2024-06-05 Classification: VIRAL PROTEIN/IMMUNE SYSTEM Ligands: NAG, SO4 |
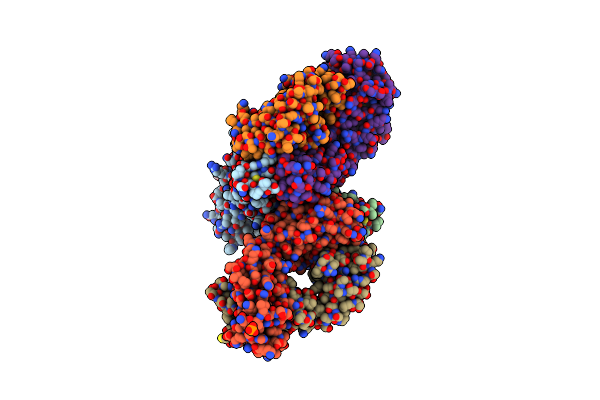 |
Organism: Severe acute respiratory syndrome coronavirus 2, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.94 Å Release Date: 2024-06-05 Classification: VIRAL PROTEIN/IMMUNE SYSTEM Ligands: CL, PO4 |
 |
Organism: Severe acute respiratory syndrome coronavirus 2, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-06-05 Classification: VIRAL PROTEIN/IMMUNE SYSTEM |
 |
Organism: Severe acute respiratory syndrome coronavirus 2, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-06-05 Classification: VIRAL PROTEIN/IMMUNE SYSTEM |
 |
Organism: Severe acute respiratory syndrome coronavirus 2, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-06-05 Classification: VIRAL PROTEIN/IMMUNE SYSTEM |
 |
Organism: Severe acute respiratory syndrome coronavirus 2, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-06-05 Classification: VIRAL PROTEIN/IMMUNE SYSTEM Ligands: NAG |
 |
Organism: Pseudomonas phage dms3
Method: X-RAY DIFFRACTION Resolution:1.33 Å Release Date: 2024-05-08 Classification: VIRAL PROTEIN |
 |
Organism: Simonsiella muelleri
Method: SOLUTION NMR Release Date: 2024-05-08 Classification: PROTEIN BINDING |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.76 Å Release Date: 2024-04-17 Classification: PEPTIDE BINDING PROTEIN/ANTIBIOTIC |
 |
Crystal Structure Of Co Dehydrogenase Mutant With Increased Affinity For Electron Mediators In High Peg Concentration
Organism: Carboxydothermus hydrogenoformans z-2901
Method: X-RAY DIFFRACTION Resolution:2.11 Å Release Date: 2024-04-17 Classification: ELECTRON TRANSPORT Ligands: SF4, FES, XCC, FE, 1PE |
 |
Crystal Structure Of Co Dehydrogenase Mutant With Increased Affinity For Electron Mediators In Low Peg Concentration
Organism: Carboxydothermus hydrogenoformans z-2901
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2024-04-17 Classification: ELECTRON TRANSPORT Ligands: SF4, FES, XCC, FE, EDO |
 |
Organism: Carboxydothermus hydrogenoformans z-2901
Method: X-RAY DIFFRACTION Resolution:2.48 Å Release Date: 2024-04-17 Classification: ELECTRON TRANSPORT Ligands: SF4, FES, XCC, S8I, EDO |
 |
Organism: Carboxydothermus hydrogenoformans z-2901
Method: X-RAY DIFFRACTION Resolution:3.11 Å Release Date: 2024-04-17 Classification: ELECTRON TRANSPORT Ligands: SF4, FES, XCC, PGE, S7L |

